Chlorine in PDB 7qn5: Cryo-Em Structure of Human Full-Length Extrasynaptic ALPHA4BETA3DELTA Gaba(A)R in Complex with Nanobody NB25
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Cryo-Em Structure of Human Full-Length Extrasynaptic ALPHA4BETA3DELTA Gaba(A)R in Complex with Nanobody NB25
(pdb code 7qn5). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 3 binding sites of Chlorine where determined in the Cryo-Em Structure of Human Full-Length Extrasynaptic ALPHA4BETA3DELTA Gaba(A)R in Complex with Nanobody NB25, PDB code: 7qn5:
Jump to Chlorine binding site number: 1; 2; 3;
In total 3 binding sites of Chlorine where determined in the Cryo-Em Structure of Human Full-Length Extrasynaptic ALPHA4BETA3DELTA Gaba(A)R in Complex with Nanobody NB25, PDB code: 7qn5:
Jump to Chlorine binding site number: 1; 2; 3;
Chlorine binding site 1 out of 3 in 7qn5
Go back to
Chlorine binding site 1 out
of 3 in the Cryo-Em Structure of Human Full-Length Extrasynaptic ALPHA4BETA3DELTA Gaba(A)R in Complex with Nanobody NB25
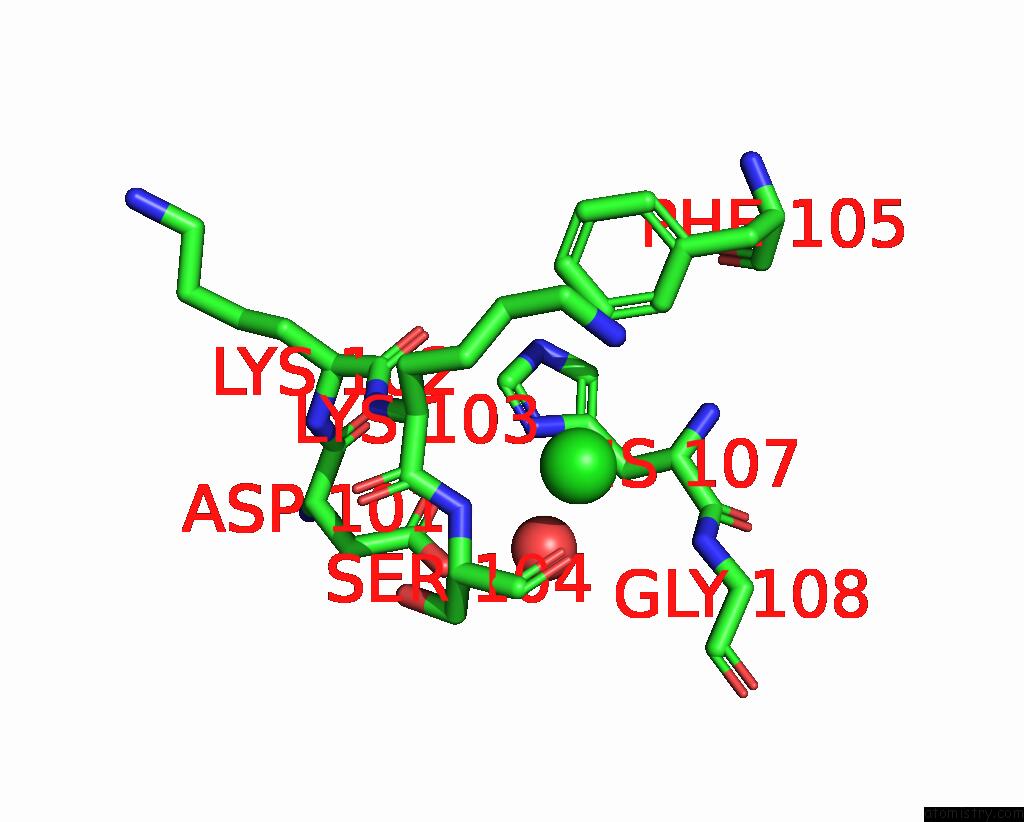
Mono view
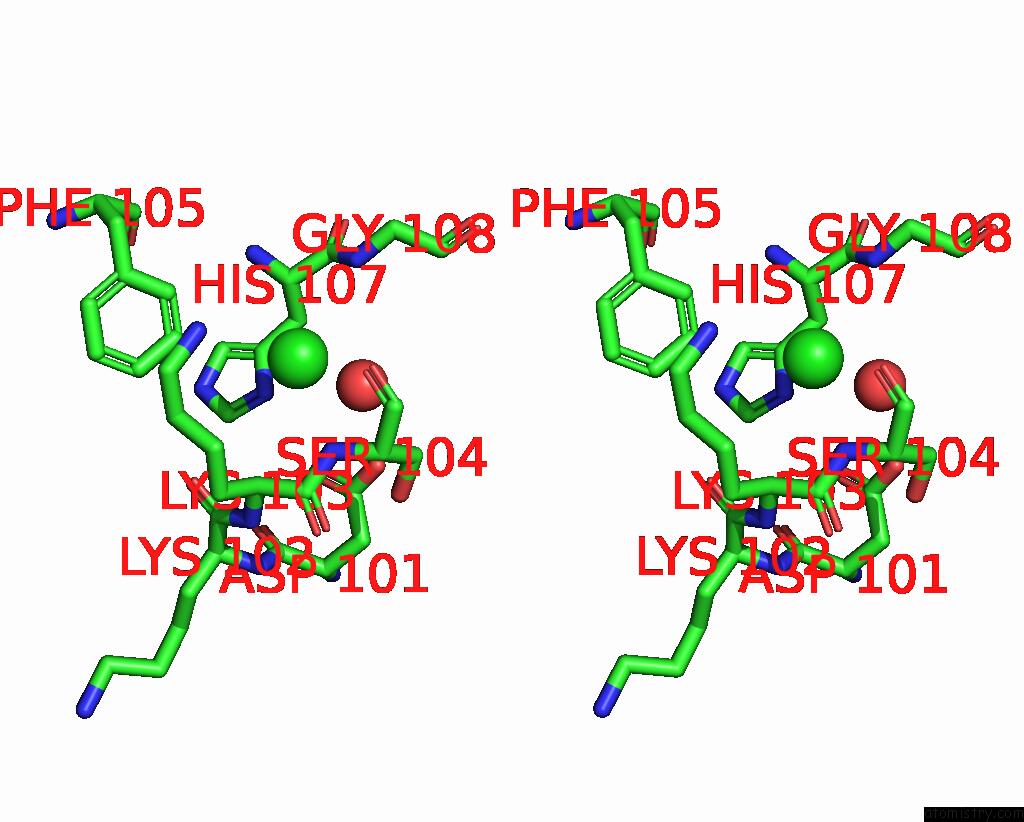
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Cryo-Em Structure of Human Full-Length Extrasynaptic ALPHA4BETA3DELTA Gaba(A)R in Complex with Nanobody NB25 within 5.0Å range:
|
Chlorine binding site 2 out of 3 in 7qn5
Go back to
Chlorine binding site 2 out
of 3 in the Cryo-Em Structure of Human Full-Length Extrasynaptic ALPHA4BETA3DELTA Gaba(A)R in Complex with Nanobody NB25
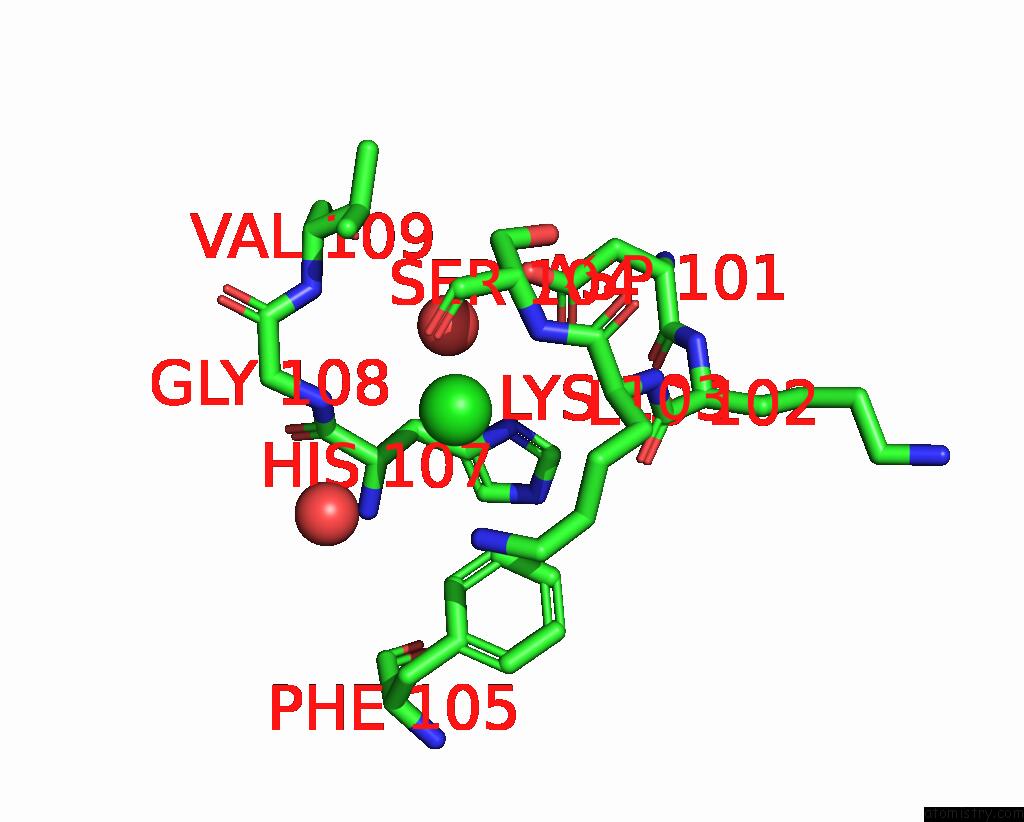
Mono view
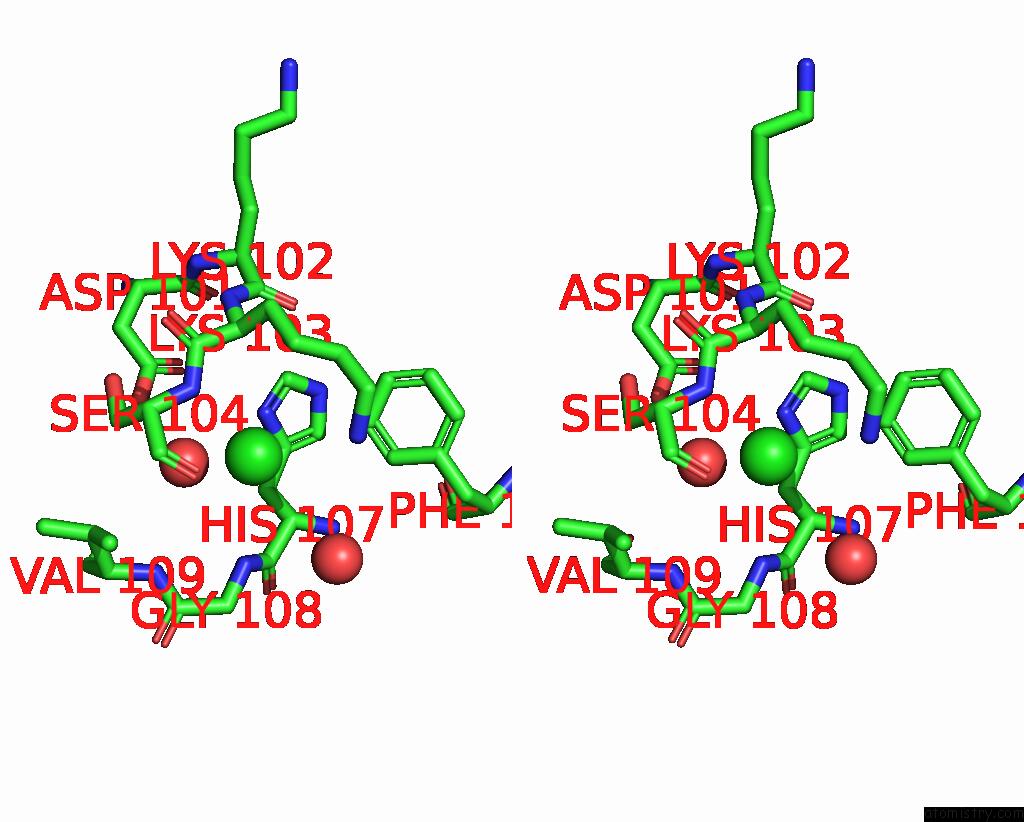
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Cryo-Em Structure of Human Full-Length Extrasynaptic ALPHA4BETA3DELTA Gaba(A)R in Complex with Nanobody NB25 within 5.0Å range:
|
Chlorine binding site 3 out of 3 in 7qn5
Go back to
Chlorine binding site 3 out
of 3 in the Cryo-Em Structure of Human Full-Length Extrasynaptic ALPHA4BETA3DELTA Gaba(A)R in Complex with Nanobody NB25

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Cryo-Em Structure of Human Full-Length Extrasynaptic ALPHA4BETA3DELTA Gaba(A)R in Complex with Nanobody NB25 within 5.0Å range:
|
Reference:
A.Sente,
R.Desai,
K.Naydenova,
T.Malinauskas,
Y.Jounaidi,
J.Miehling,
X.Zhou,
S.Masiulis,
S.W.Hardwick,
D.Y.Chirgadze,
K.W.Miller,
A.R.Aricescu.
Differential Assembly Diversifies Gaba A Receptor Structures and Signalling. Nature V. 604 190 2022.
ISSN: ESSN 1476-4687
PubMed: 35355020
DOI: 10.1038/S41586-022-04517-3
Page generated: Tue Jul 30 03:17:32 2024
ISSN: ESSN 1476-4687
PubMed: 35355020
DOI: 10.1038/S41586-022-04517-3
Last articles
Zn in 9JYWZn in 9IR4
Zn in 9IR3
Zn in 9GMX
Zn in 9GMW
Zn in 9JEJ
Zn in 9ERF
Zn in 9ERE
Zn in 9EGV
Zn in 9EGW