Chlorine in PDB 8cwg: 200US Temperature-Jump (DARK1) Xfel Structure of Lysozyme Bound to N, N'-Diacetylchitobiose
Enzymatic activity of 200US Temperature-Jump (DARK1) Xfel Structure of Lysozyme Bound to N, N'-Diacetylchitobiose
All present enzymatic activity of 200US Temperature-Jump (DARK1) Xfel Structure of Lysozyme Bound to N, N'-Diacetylchitobiose:
3.2.1.17;
3.2.1.17;
Protein crystallography data
The structure of 200US Temperature-Jump (DARK1) Xfel Structure of Lysozyme Bound to N, N'-Diacetylchitobiose, PDB code: 8cwg
was solved by
A.M.Wolff,
M.C.Thompson,
J.S.Fraser,
E.Nango,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 30.74 / 1.50 |
| Space group | P 43 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 77.061, 77.061, 37.223, 90, 90, 90 |
| R / Rfree (%) | 12.9 / 16.7 |
Other elements in 8cwg:
The structure of 200US Temperature-Jump (DARK1) Xfel Structure of Lysozyme Bound to N, N'-Diacetylchitobiose also contains other interesting chemical elements:
| Sodium | (Na) | 2 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the 200US Temperature-Jump (DARK1) Xfel Structure of Lysozyme Bound to N, N'-Diacetylchitobiose
(pdb code 8cwg). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 5 binding sites of Chlorine where determined in the 200US Temperature-Jump (DARK1) Xfel Structure of Lysozyme Bound to N, N'-Diacetylchitobiose, PDB code: 8cwg:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5;
In total 5 binding sites of Chlorine where determined in the 200US Temperature-Jump (DARK1) Xfel Structure of Lysozyme Bound to N, N'-Diacetylchitobiose, PDB code: 8cwg:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5;
Chlorine binding site 1 out of 5 in 8cwg
Go back to
Chlorine binding site 1 out
of 5 in the 200US Temperature-Jump (DARK1) Xfel Structure of Lysozyme Bound to N, N'-Diacetylchitobiose
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of 200US Temperature-Jump (DARK1) Xfel Structure of Lysozyme Bound to N, N'-Diacetylchitobiose within 5.0Å range:
|
Chlorine binding site 2 out of 5 in 8cwg
Go back to
Chlorine binding site 2 out
of 5 in the 200US Temperature-Jump (DARK1) Xfel Structure of Lysozyme Bound to N, N'-Diacetylchitobiose
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of 200US Temperature-Jump (DARK1) Xfel Structure of Lysozyme Bound to N, N'-Diacetylchitobiose within 5.0Å range:
|
Chlorine binding site 3 out of 5 in 8cwg
Go back to
Chlorine binding site 3 out
of 5 in the 200US Temperature-Jump (DARK1) Xfel Structure of Lysozyme Bound to N, N'-Diacetylchitobiose

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of 200US Temperature-Jump (DARK1) Xfel Structure of Lysozyme Bound to N, N'-Diacetylchitobiose within 5.0Å range:
|
Chlorine binding site 4 out of 5 in 8cwg
Go back to
Chlorine binding site 4 out
of 5 in the 200US Temperature-Jump (DARK1) Xfel Structure of Lysozyme Bound to N, N'-Diacetylchitobiose
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of 200US Temperature-Jump (DARK1) Xfel Structure of Lysozyme Bound to N, N'-Diacetylchitobiose within 5.0Å range:
|
Chlorine binding site 5 out of 5 in 8cwg
Go back to
Chlorine binding site 5 out
of 5 in the 200US Temperature-Jump (DARK1) Xfel Structure of Lysozyme Bound to N, N'-Diacetylchitobiose
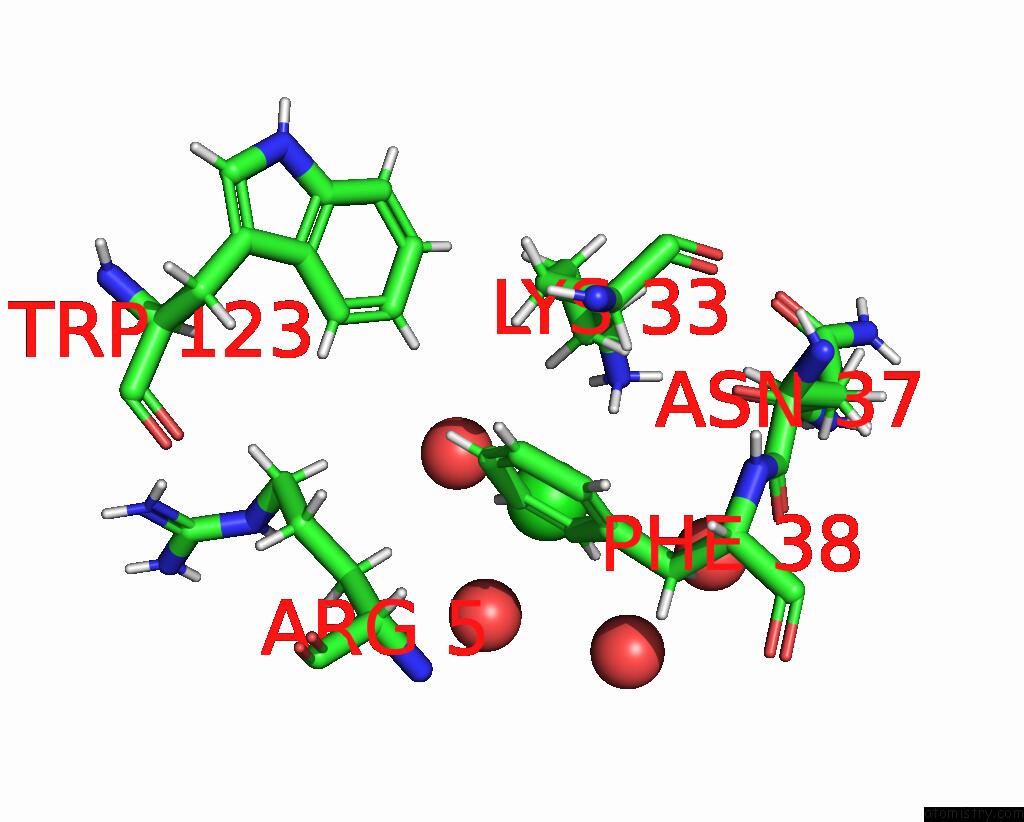
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of 200US Temperature-Jump (DARK1) Xfel Structure of Lysozyme Bound to N, N'-Diacetylchitobiose within 5.0Å range:
|
Reference:
A.M.Wolff,
E.Nango,
I.D.Young,
A.S.Brewster,
M.Kubo,
T.Nomura,
M.Sugahara,
S.Owada,
B.A.Barad,
K.Ito,
A.Bhowmick,
S.Carbajo,
T.Hino,
J.M.Holton,
D.Im,
L.J.O'riordan,
T.Tanaka,
R.Tanaka,
R.G.Sierra,
F.Yumoto,
K.Tono,
S.Iwata,
N.K.Sauter,
J.S.Fraser,
M.C.Thompson.
Mapping Protein Dynamics at High-Resolution with Temperature-Jump X-Ray Crystallography Biorxiv 2022.
DOI: 10.1101/2022.06.10.495662
Page generated: Tue Jul 30 08:13:49 2024
DOI: 10.1101/2022.06.10.495662
Last articles
Zn in 9JYWZn in 9IR4
Zn in 9IR3
Zn in 9GMX
Zn in 9GMW
Zn in 9JEJ
Zn in 9ERF
Zn in 9ERE
Zn in 9EGV
Zn in 9EGW