Chlorine in PDB 8dv3: Crystal Structure of Human CD1B Presenting Phosphatidylinositol C34:1
Protein crystallography data
The structure of Crystal Structure of Human CD1B Presenting Phosphatidylinositol C34:1, PDB code: 8dv3
was solved by
R.Farquhar,
J.Rossjohn,
A.Shahine,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 46.58 / 1.90 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 57.816, 78.628, 92.725, 90, 90, 90 |
| R / Rfree (%) | 19.2 / 25.3 |
Other elements in 8dv3:
The structure of Crystal Structure of Human CD1B Presenting Phosphatidylinositol C34:1 also contains other interesting chemical elements:
| Sodium | (Na) | 2 atoms |
| Iodine | (I) | 2 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of Human CD1B Presenting Phosphatidylinositol C34:1
(pdb code 8dv3). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 6 binding sites of Chlorine where determined in the Crystal Structure of Human CD1B Presenting Phosphatidylinositol C34:1, PDB code: 8dv3:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Chlorine where determined in the Crystal Structure of Human CD1B Presenting Phosphatidylinositol C34:1, PDB code: 8dv3:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6;
Chlorine binding site 1 out of 6 in 8dv3
Go back to
Chlorine binding site 1 out
of 6 in the Crystal Structure of Human CD1B Presenting Phosphatidylinositol C34:1
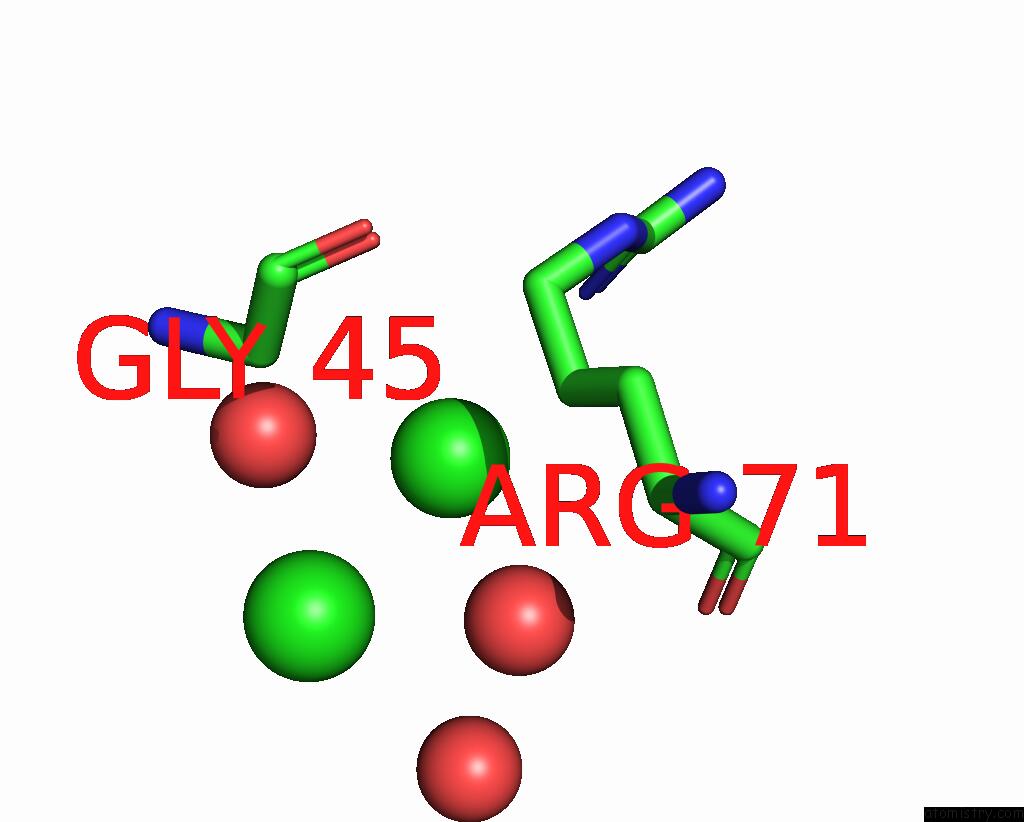
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of Human CD1B Presenting Phosphatidylinositol C34:1 within 5.0Å range:
|
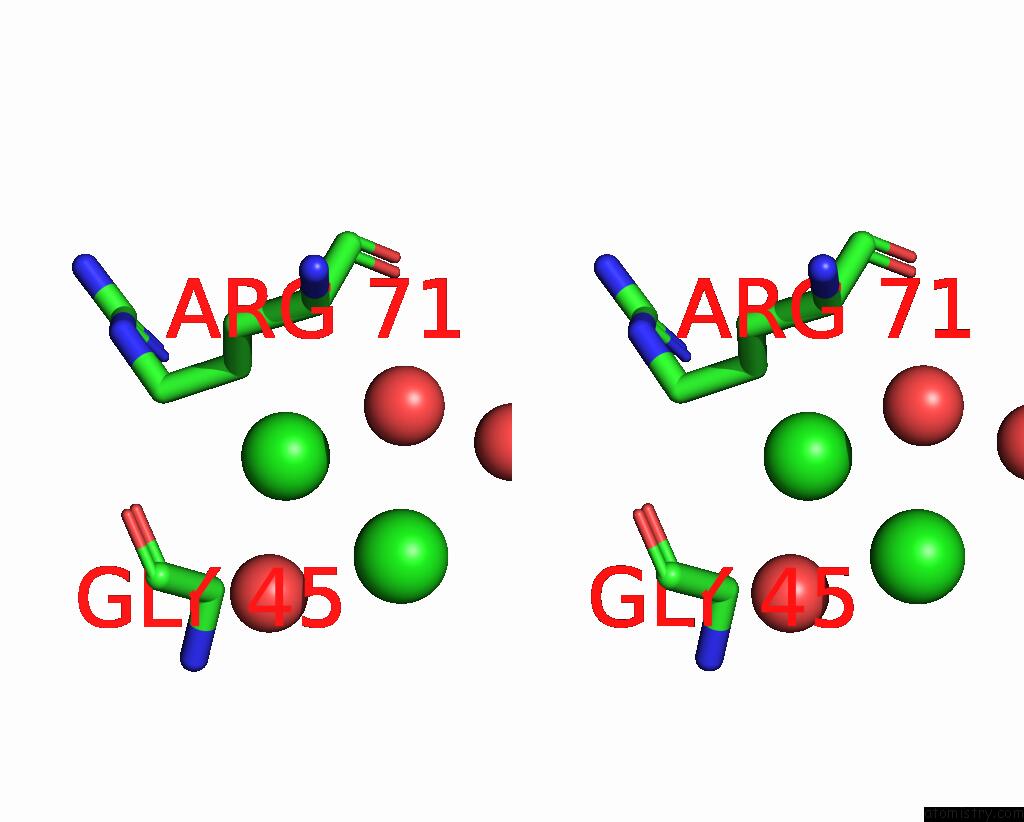
Chlorine binding site 2 out of 6 in 8dv3
Go back to
Chlorine binding site 2 out
of 6 in the Crystal Structure of Human CD1B Presenting Phosphatidylinositol C34:1
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of Human CD1B Presenting Phosphatidylinositol C34:1 within 5.0Å range:
|
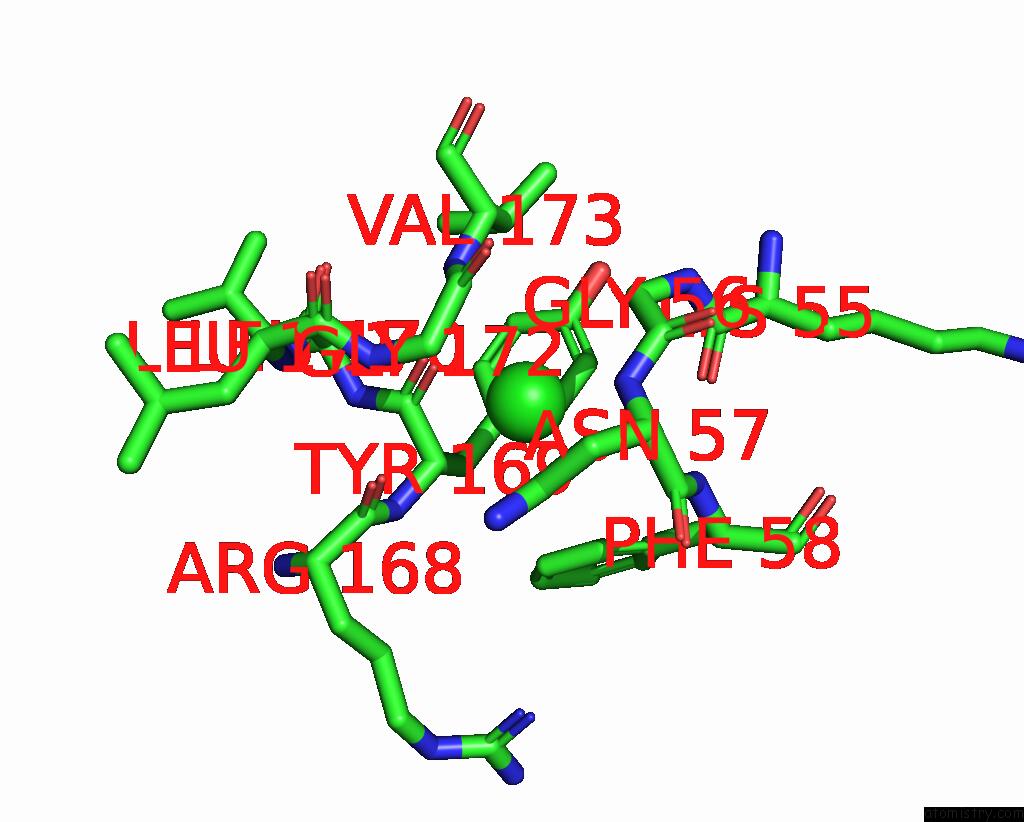
Chlorine binding site 3 out of 6 in 8dv3
Go back to
Chlorine binding site 3 out
of 6 in the Crystal Structure of Human CD1B Presenting Phosphatidylinositol C34:1
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Crystal Structure of Human CD1B Presenting Phosphatidylinositol C34:1 within 5.0Å range:
|
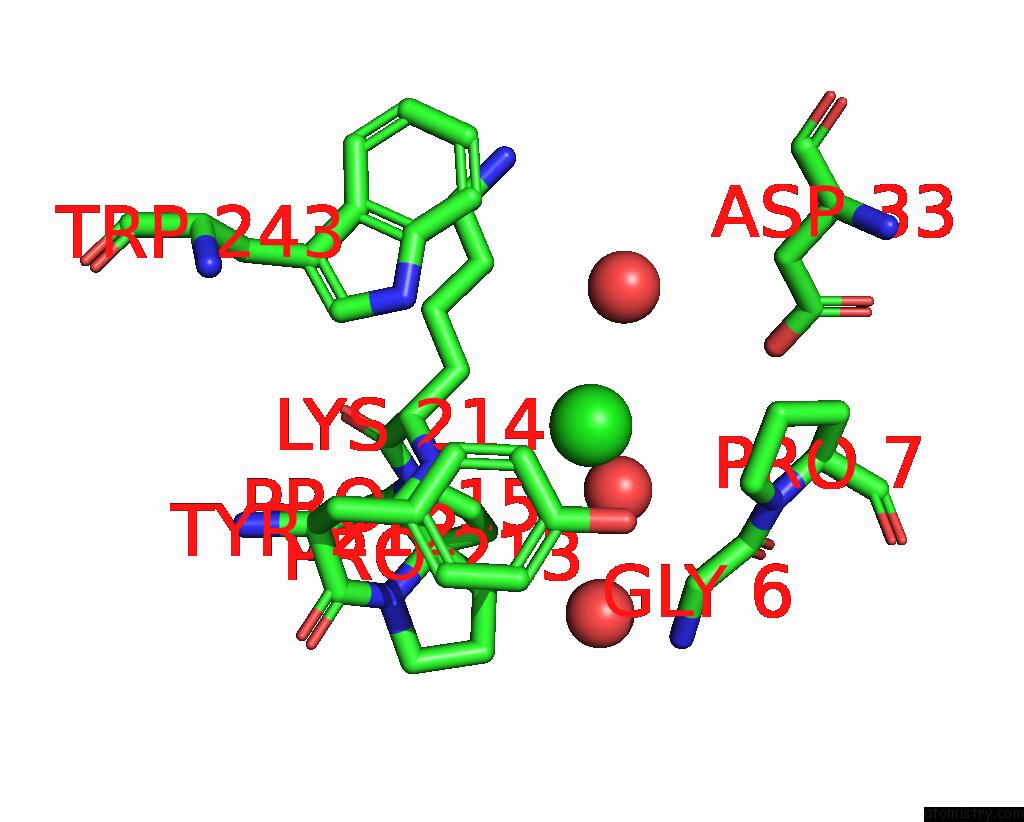
Chlorine binding site 4 out of 6 in 8dv3
Go back to
Chlorine binding site 4 out
of 6 in the Crystal Structure of Human CD1B Presenting Phosphatidylinositol C34:1
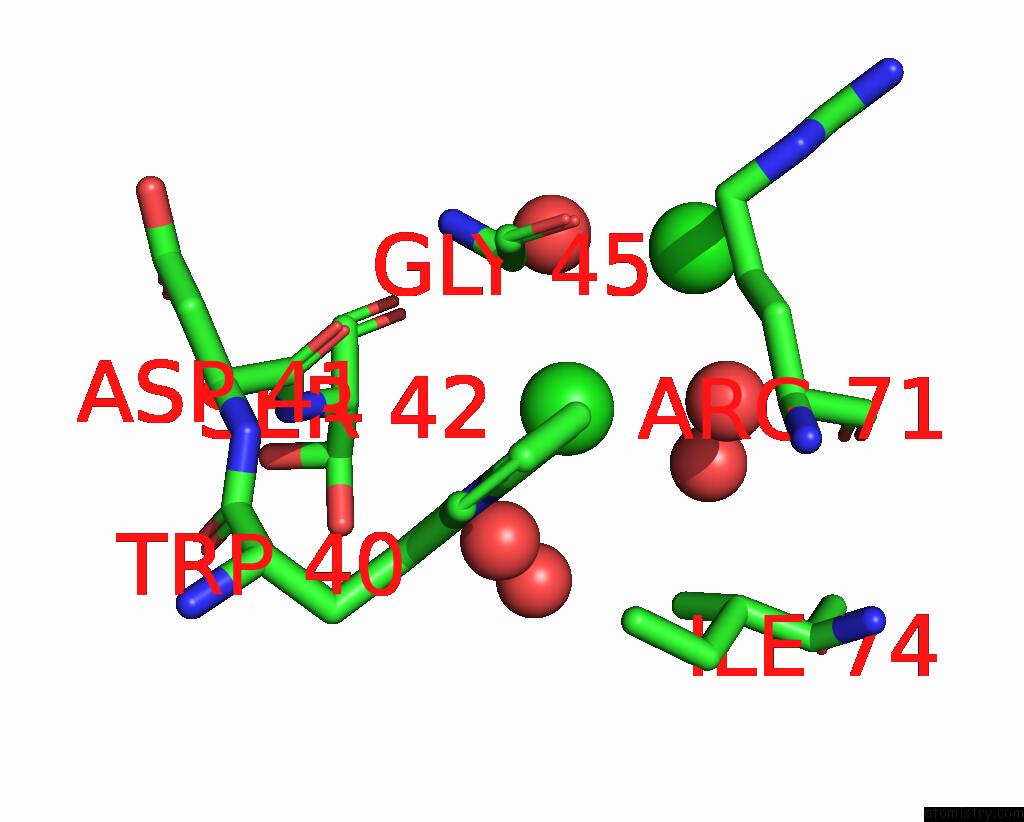
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Crystal Structure of Human CD1B Presenting Phosphatidylinositol C34:1 within 5.0Å range:
|
Chlorine binding site 5 out of 6 in 8dv3
Go back to
Chlorine binding site 5 out
of 6 in the Crystal Structure of Human CD1B Presenting Phosphatidylinositol C34:1

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Crystal Structure of Human CD1B Presenting Phosphatidylinositol C34:1 within 5.0Å range:
|
Chlorine binding site 6 out of 6 in 8dv3
Go back to
Chlorine binding site 6 out
of 6 in the Crystal Structure of Human CD1B Presenting Phosphatidylinositol C34:1
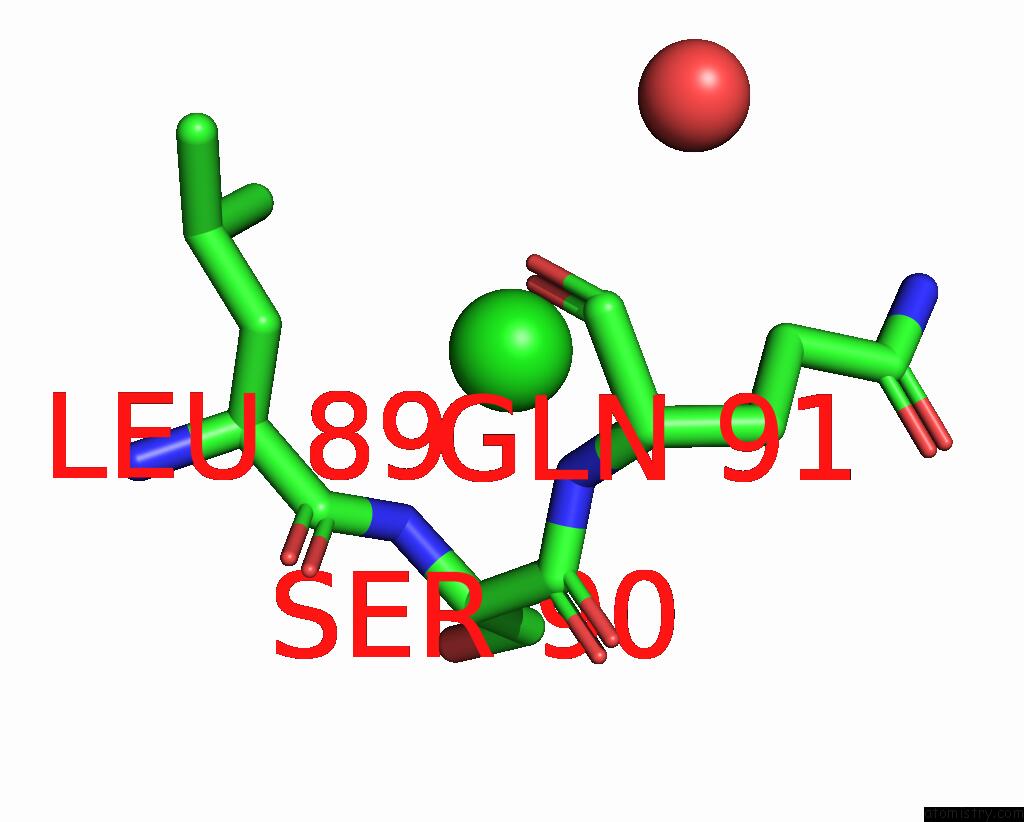
Mono view
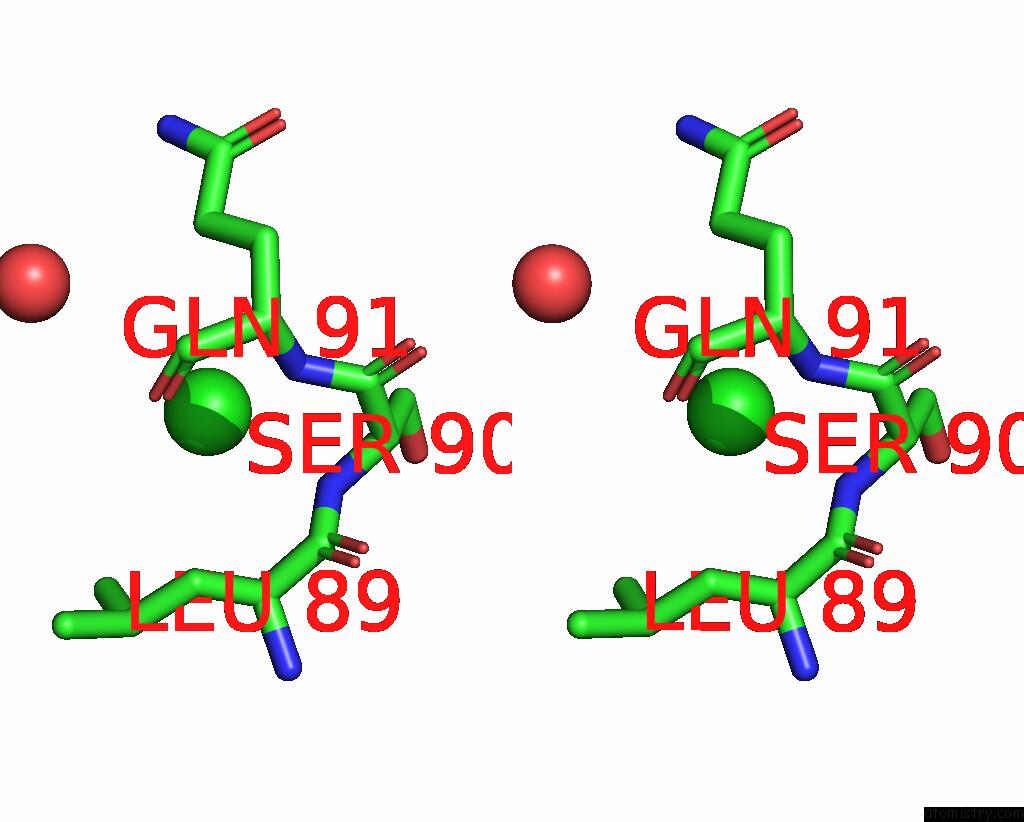
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 6 of Crystal Structure of Human CD1B Presenting Phosphatidylinositol C34:1 within 5.0Å range:
|
Reference:
R.Farquhar,
I.Van Rhijn,
D.B.Moody,
J.Rossjohn,
A.Shahine.
Alpha Beta T-Cell Receptor Recognition of Self-Phosphatidylinositol Presented By CD1B. J.Biol.Chem. V. 299 02849 2023.
ISSN: ESSN 1083-351X
PubMed: 36587766
DOI: 10.1016/J.JBC.2022.102849
Page generated: Tue Jul 30 08:54:09 2024
ISSN: ESSN 1083-351X
PubMed: 36587766
DOI: 10.1016/J.JBC.2022.102849
Last articles
Zn in 9J0NZn in 9J0O
Zn in 9J0P
Zn in 9FJX
Zn in 9EKB
Zn in 9C0F
Zn in 9CAH
Zn in 9CH0
Zn in 9CH3
Zn in 9CH1