Chlorine in PDB 8fp9: GLUA2 Flip Q Isoform of Ampa Receptor in Complex with Gain-of-Function Tarp Gamma-2, with 10MM CACL2, 150MM Nacl, 1MM MGCL2, 330UM Ctz, and 100MM Glutamate (Open-Canamg)
Other elements in 8fp9:
The structure of GLUA2 Flip Q Isoform of Ampa Receptor in Complex with Gain-of-Function Tarp Gamma-2, with 10MM CACL2, 150MM Nacl, 1MM MGCL2, 330UM Ctz, and 100MM Glutamate (Open-Canamg) also contains other interesting chemical elements:
| Calcium | (Ca) | 1 atom |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the GLUA2 Flip Q Isoform of Ampa Receptor in Complex with Gain-of-Function Tarp Gamma-2, with 10MM CACL2, 150MM Nacl, 1MM MGCL2, 330UM Ctz, and 100MM Glutamate (Open-Canamg)
(pdb code 8fp9). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 4 binding sites of Chlorine where determined in the GLUA2 Flip Q Isoform of Ampa Receptor in Complex with Gain-of-Function Tarp Gamma-2, with 10MM CACL2, 150MM Nacl, 1MM MGCL2, 330UM Ctz, and 100MM Glutamate (Open-Canamg), PDB code: 8fp9:
Jump to Chlorine binding site number: 1; 2; 3; 4;
In total 4 binding sites of Chlorine where determined in the GLUA2 Flip Q Isoform of Ampa Receptor in Complex with Gain-of-Function Tarp Gamma-2, with 10MM CACL2, 150MM Nacl, 1MM MGCL2, 330UM Ctz, and 100MM Glutamate (Open-Canamg), PDB code: 8fp9:
Jump to Chlorine binding site number: 1; 2; 3; 4;
Chlorine binding site 1 out of 4 in 8fp9
Go back to
Chlorine binding site 1 out
of 4 in the GLUA2 Flip Q Isoform of Ampa Receptor in Complex with Gain-of-Function Tarp Gamma-2, with 10MM CACL2, 150MM Nacl, 1MM MGCL2, 330UM Ctz, and 100MM Glutamate (Open-Canamg)
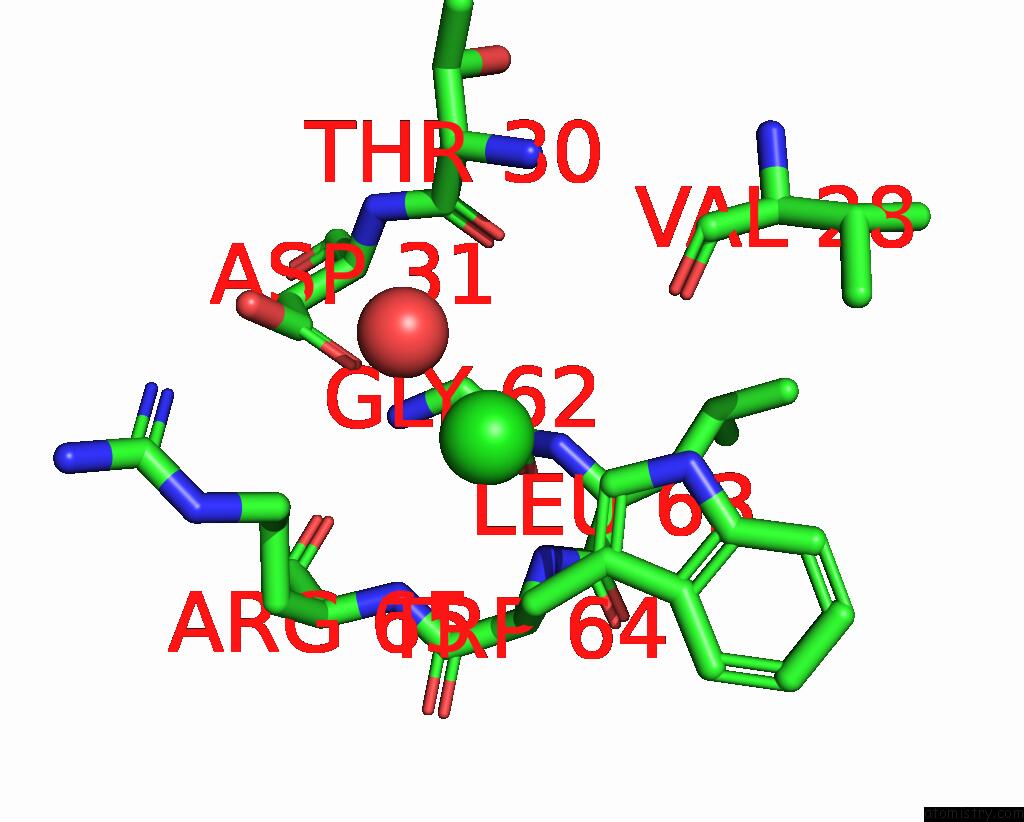
Mono view
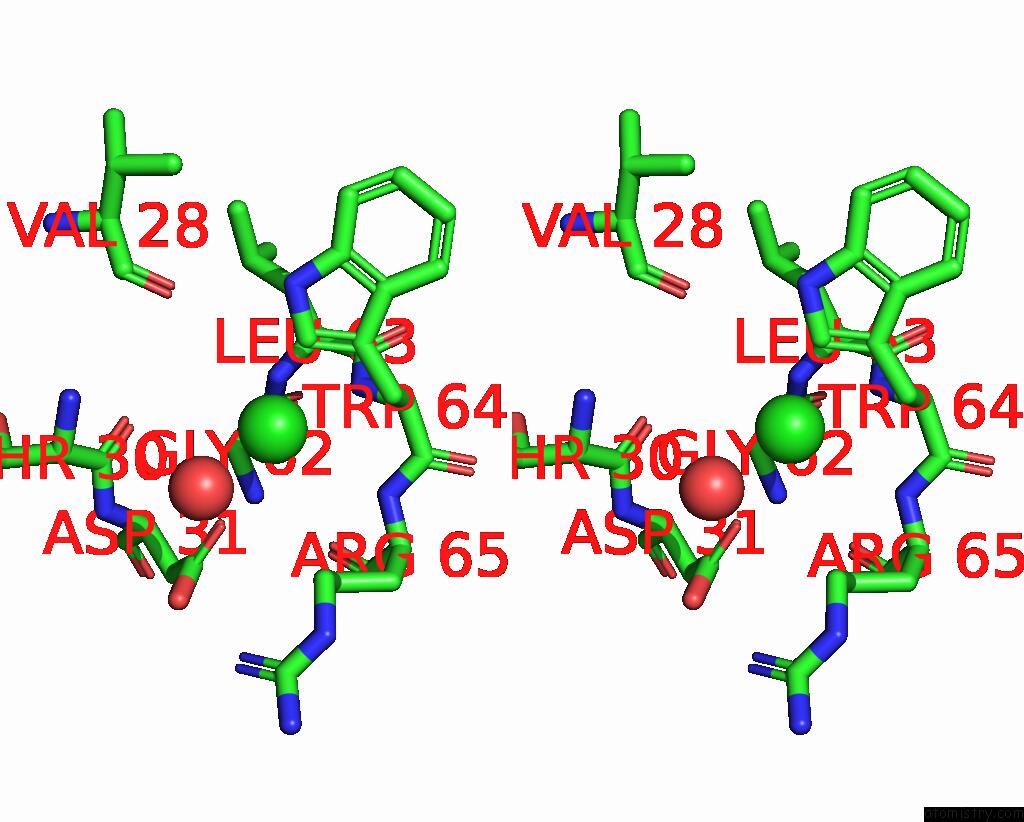
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of GLUA2 Flip Q Isoform of Ampa Receptor in Complex with Gain-of-Function Tarp Gamma-2, with 10MM CACL2, 150MM Nacl, 1MM MGCL2, 330UM Ctz, and 100MM Glutamate (Open-Canamg) within 5.0Å range:
|
Chlorine binding site 2 out of 4 in 8fp9
Go back to
Chlorine binding site 2 out
of 4 in the GLUA2 Flip Q Isoform of Ampa Receptor in Complex with Gain-of-Function Tarp Gamma-2, with 10MM CACL2, 150MM Nacl, 1MM MGCL2, 330UM Ctz, and 100MM Glutamate (Open-Canamg)
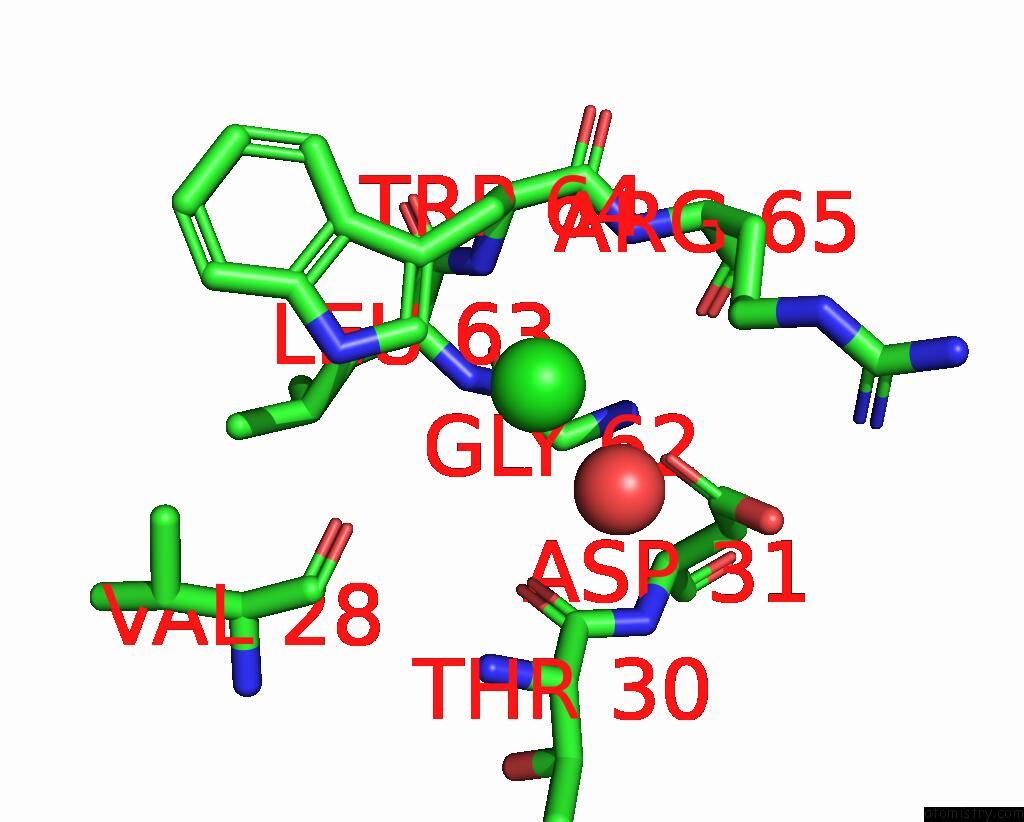
Mono view
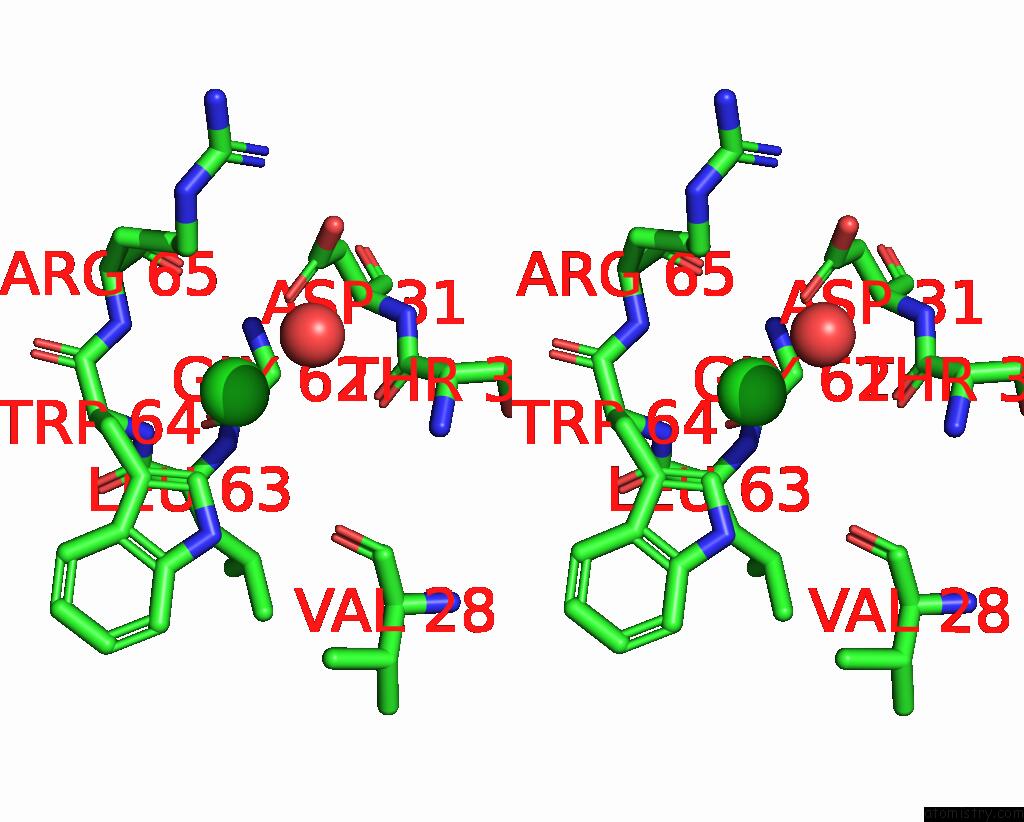
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of GLUA2 Flip Q Isoform of Ampa Receptor in Complex with Gain-of-Function Tarp Gamma-2, with 10MM CACL2, 150MM Nacl, 1MM MGCL2, 330UM Ctz, and 100MM Glutamate (Open-Canamg) within 5.0Å range:
|
Chlorine binding site 3 out of 4 in 8fp9
Go back to
Chlorine binding site 3 out
of 4 in the GLUA2 Flip Q Isoform of Ampa Receptor in Complex with Gain-of-Function Tarp Gamma-2, with 10MM CACL2, 150MM Nacl, 1MM MGCL2, 330UM Ctz, and 100MM Glutamate (Open-Canamg)
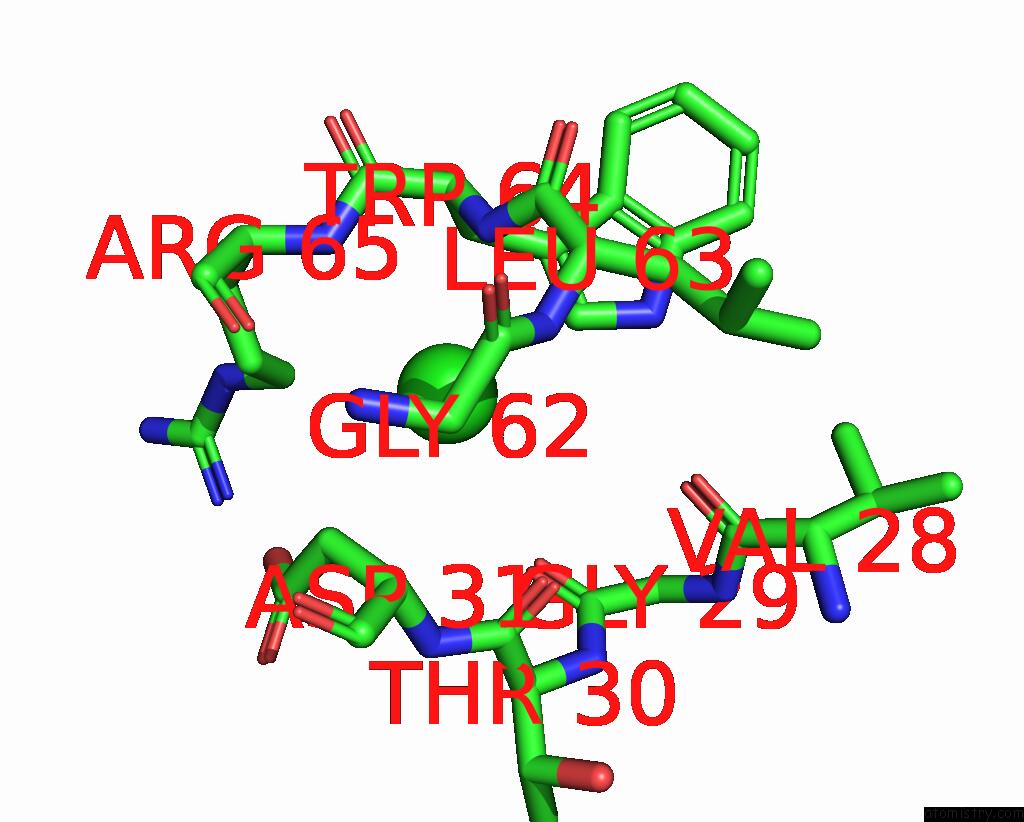
Mono view
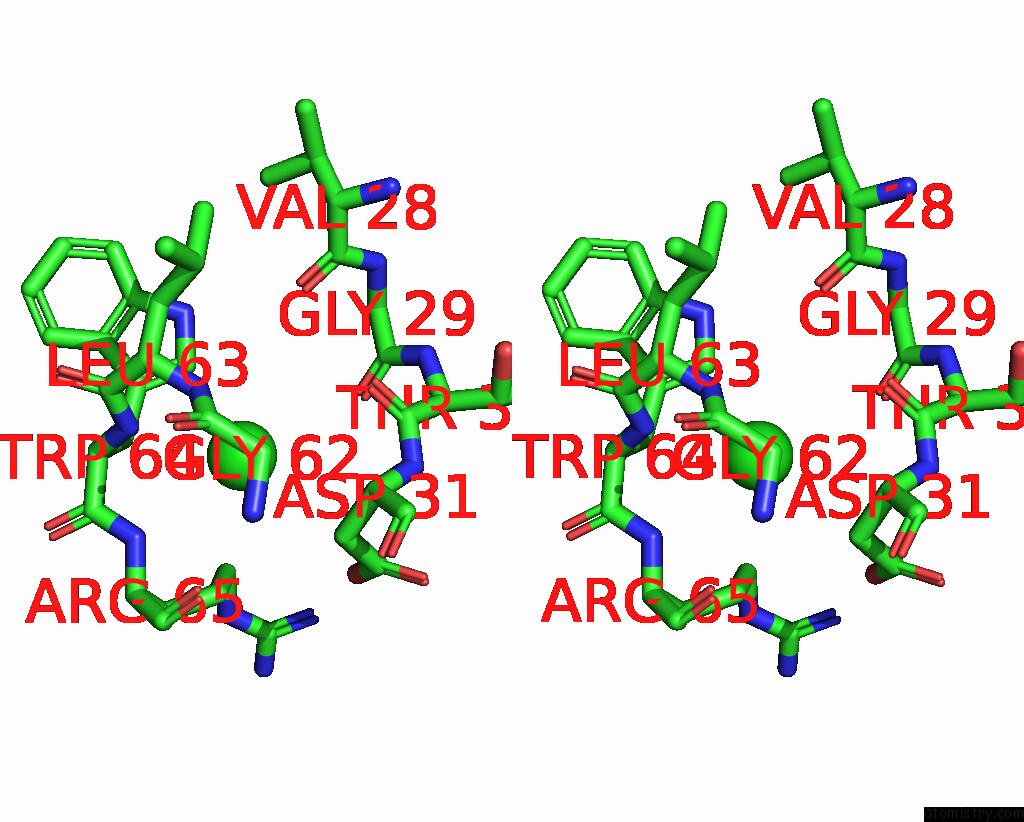
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of GLUA2 Flip Q Isoform of Ampa Receptor in Complex with Gain-of-Function Tarp Gamma-2, with 10MM CACL2, 150MM Nacl, 1MM MGCL2, 330UM Ctz, and 100MM Glutamate (Open-Canamg) within 5.0Å range:
|
Chlorine binding site 4 out of 4 in 8fp9
Go back to
Chlorine binding site 4 out
of 4 in the GLUA2 Flip Q Isoform of Ampa Receptor in Complex with Gain-of-Function Tarp Gamma-2, with 10MM CACL2, 150MM Nacl, 1MM MGCL2, 330UM Ctz, and 100MM Glutamate (Open-Canamg)
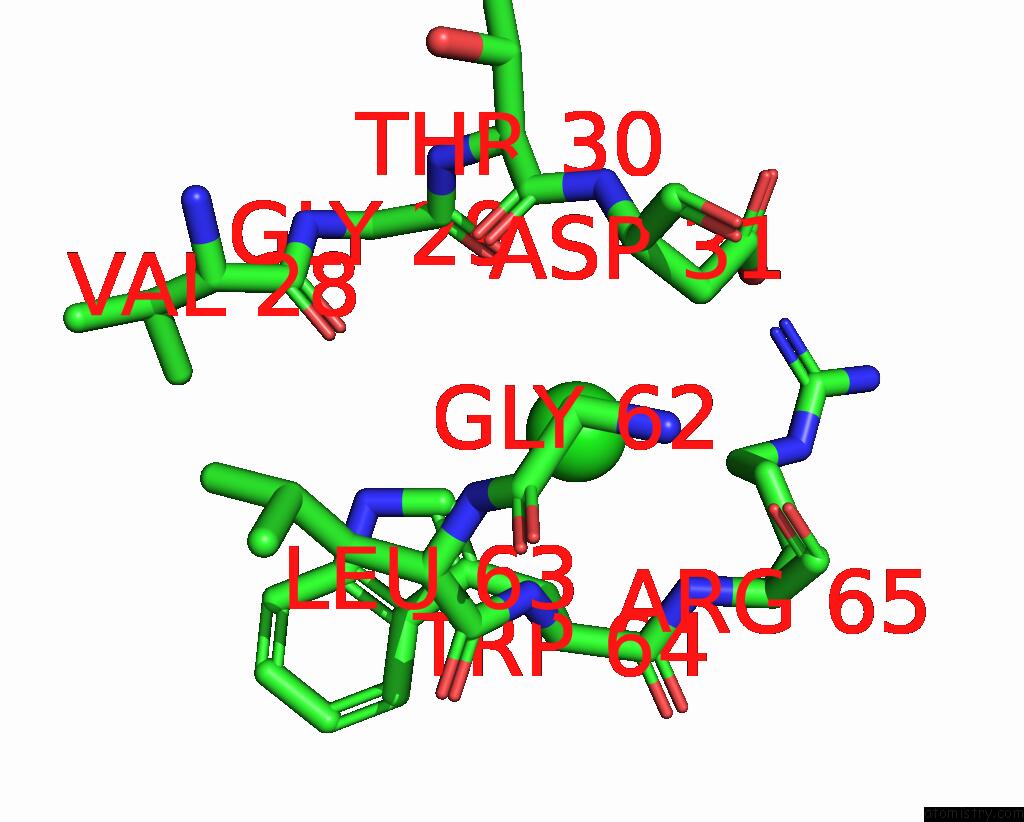
Mono view
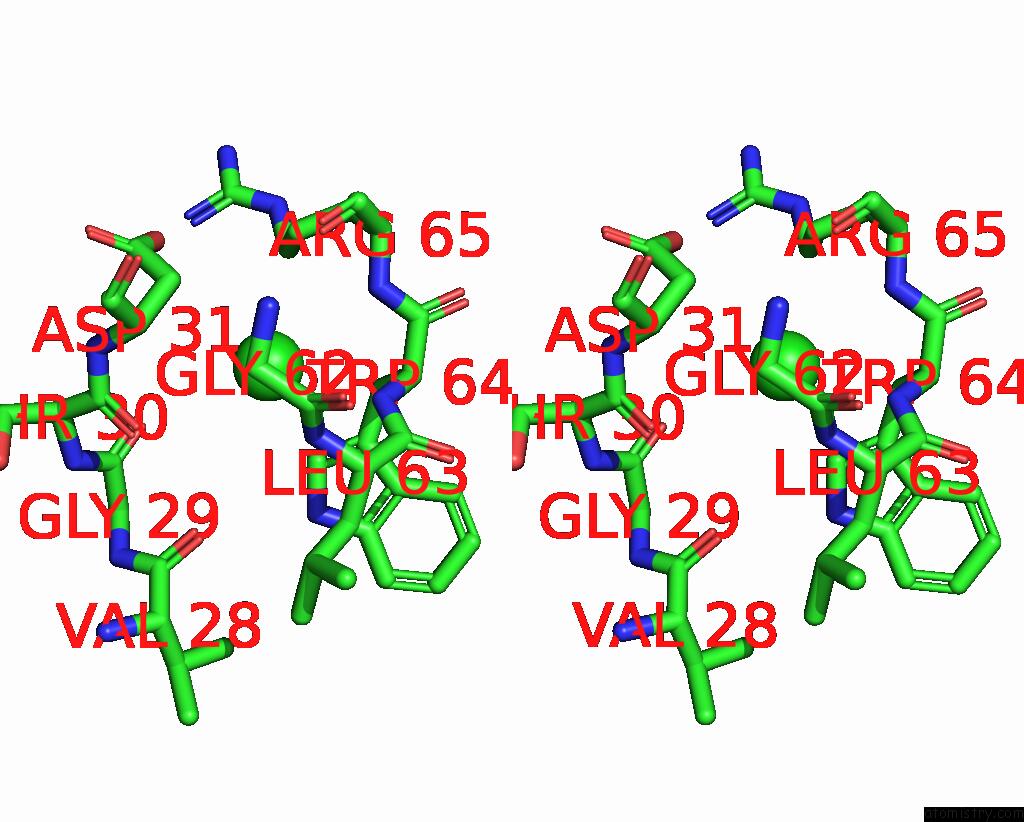
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of GLUA2 Flip Q Isoform of Ampa Receptor in Complex with Gain-of-Function Tarp Gamma-2, with 10MM CACL2, 150MM Nacl, 1MM MGCL2, 330UM Ctz, and 100MM Glutamate (Open-Canamg) within 5.0Å range:
|
Reference:
T.Nakagawa,
X.Wang,
F.J.Miguez-Cabello,
D.Bowie.
The Open Gate of the Ampa Receptor Forms A CA2+ Binding Site Critical in Regulating Ion Transport Nat.Struct.Mol.Biol. 2024.
ISSN: ESSN 1545-9985
DOI: 10.1038/S41594-024-01228-3
Page generated: Tue Jul 30 09:39:59 2024
ISSN: ESSN 1545-9985
DOI: 10.1038/S41594-024-01228-3
Last articles
Zn in 9JYWZn in 9IR4
Zn in 9IR3
Zn in 9GMX
Zn in 9GMW
Zn in 9JEJ
Zn in 9ERF
Zn in 9ERE
Zn in 9EGV
Zn in 9EGW