Chlorine in PDB 8yak: Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase
Other elements in 8yak:
The structure of Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase also contains other interesting chemical elements:
| Zinc | (Zn) | 16 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase
(pdb code 8yak). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 8 binding sites of Chlorine where determined in the Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase, PDB code: 8yak:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
In total 8 binding sites of Chlorine where determined in the Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase, PDB code: 8yak:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
Chlorine binding site 1 out of 8 in 8yak
Go back to
Chlorine binding site 1 out
of 8 in the Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase
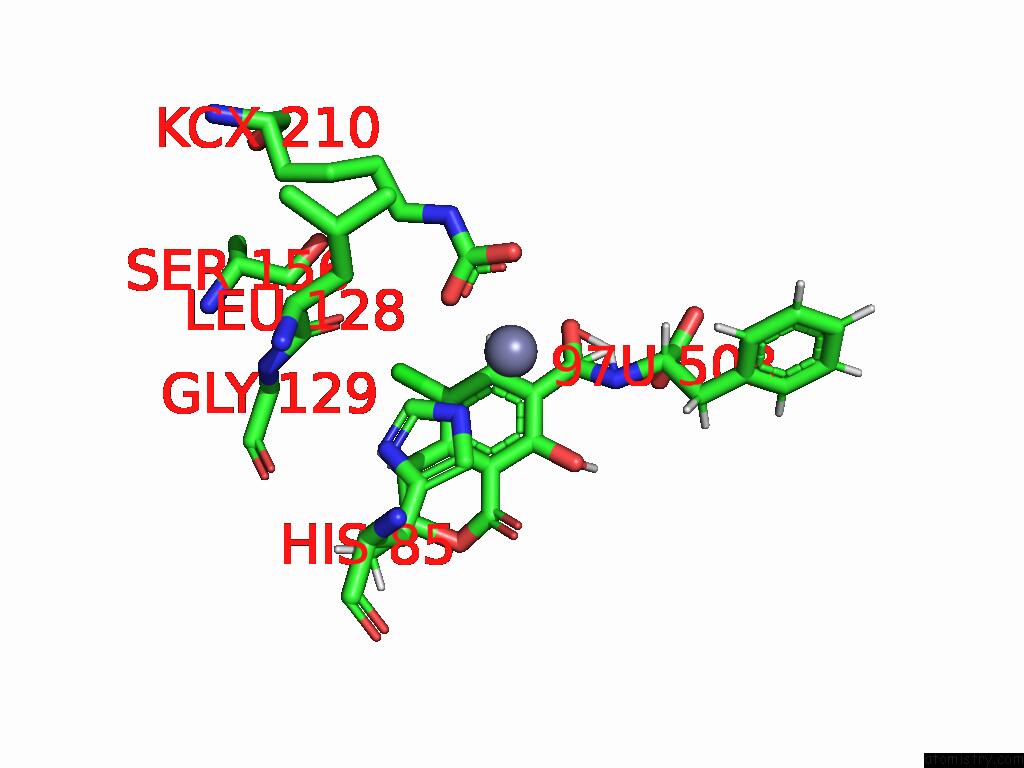
Mono view
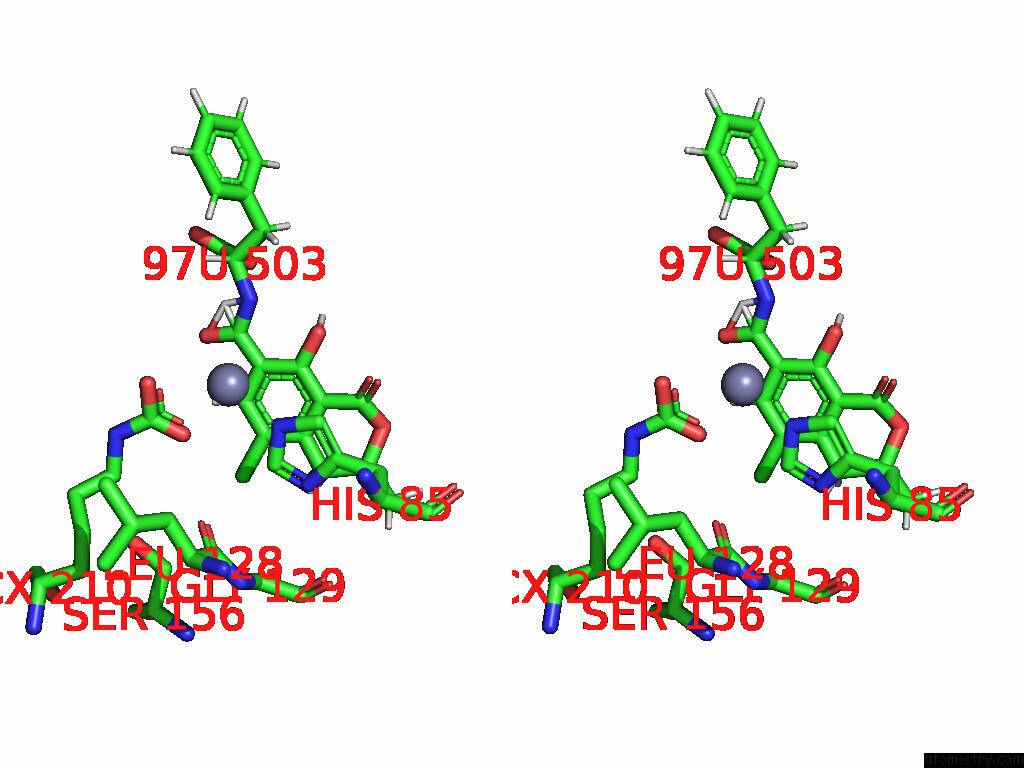
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase within 5.0Å range:
|
Chlorine binding site 2 out of 8 in 8yak
Go back to
Chlorine binding site 2 out
of 8 in the Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase
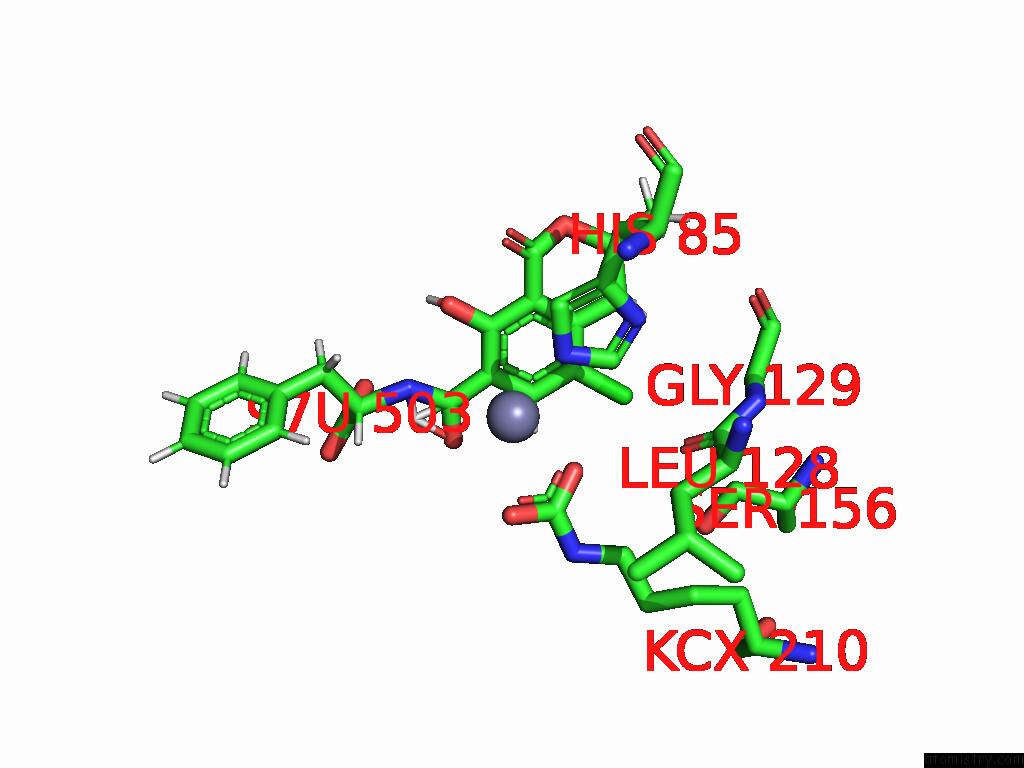
Mono view
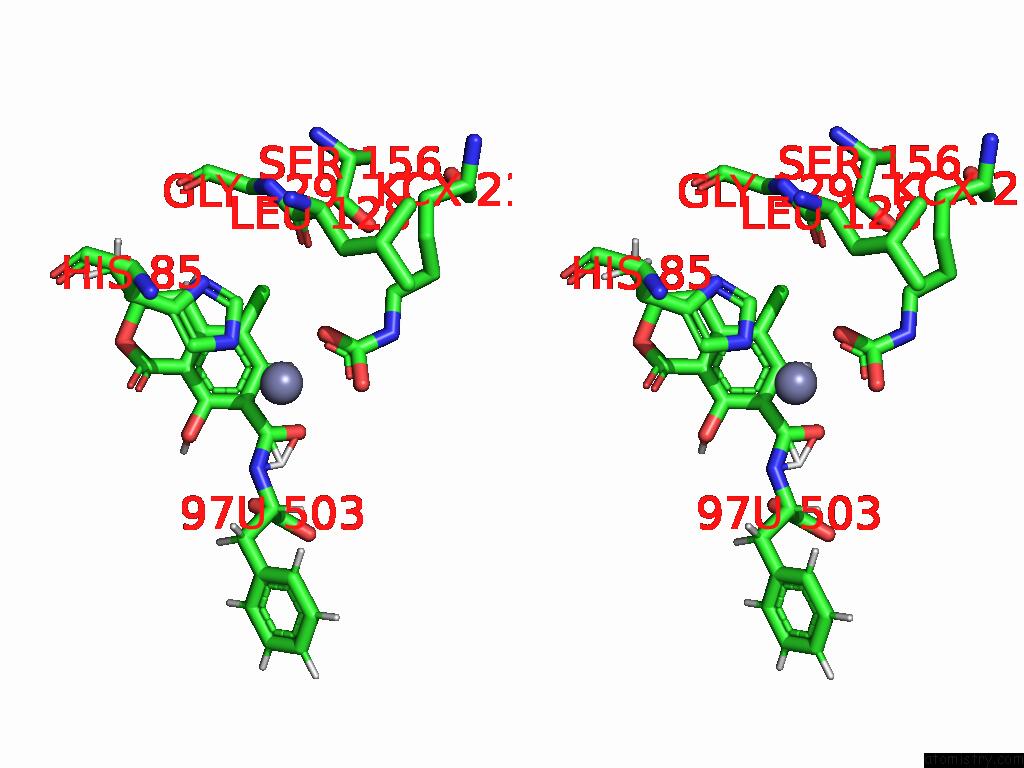
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase within 5.0Å range:
|
Chlorine binding site 3 out of 8 in 8yak
Go back to
Chlorine binding site 3 out
of 8 in the Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase
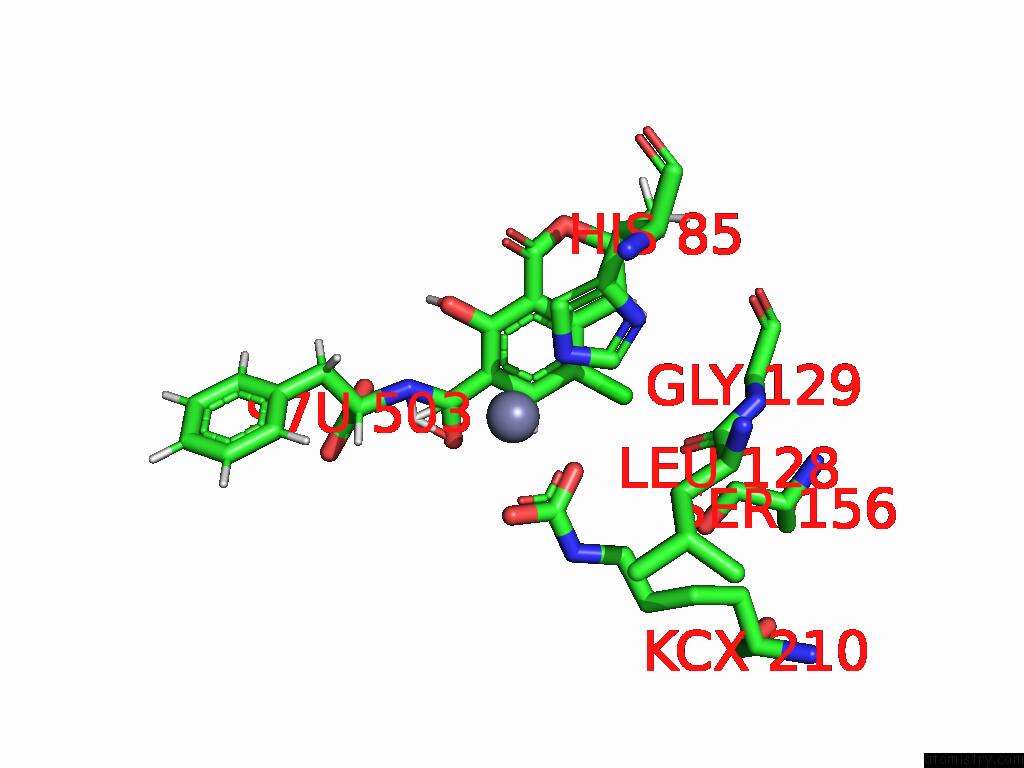
Mono view
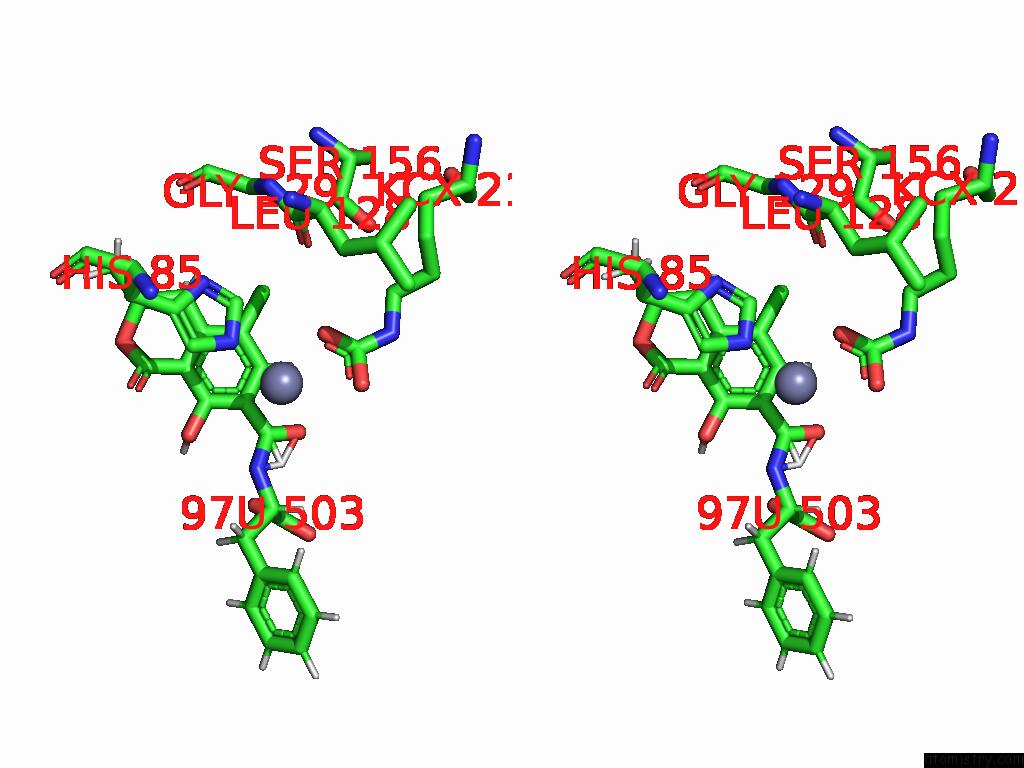
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase within 5.0Å range:
|
Chlorine binding site 4 out of 8 in 8yak
Go back to
Chlorine binding site 4 out
of 8 in the Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase
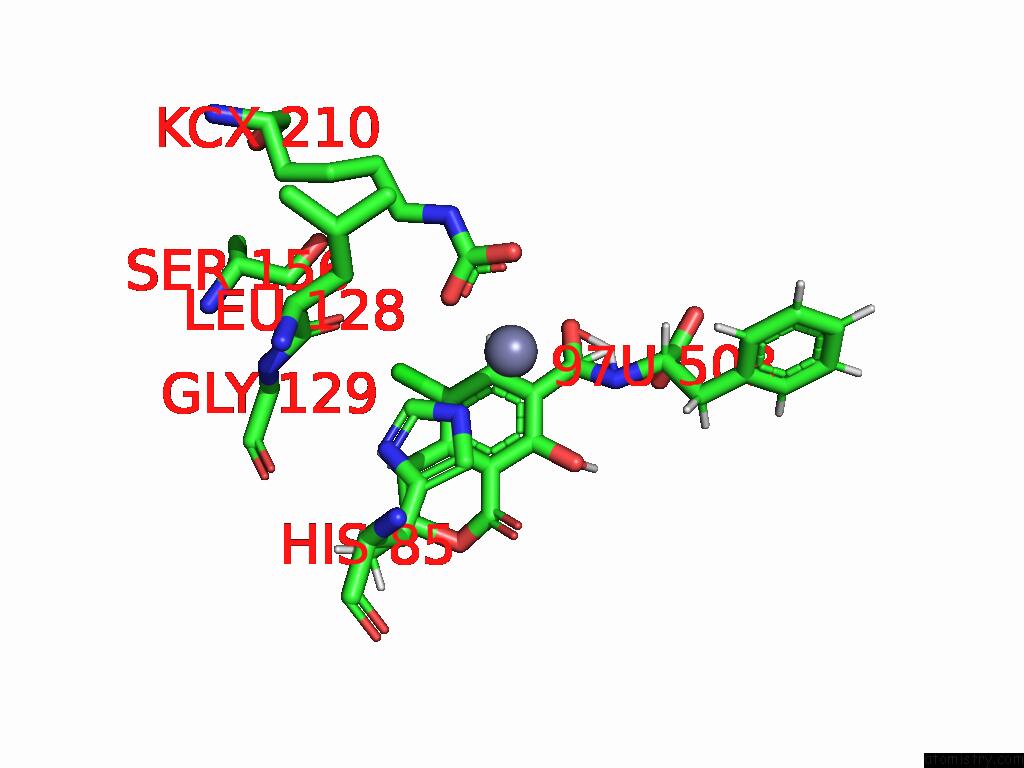
Mono view
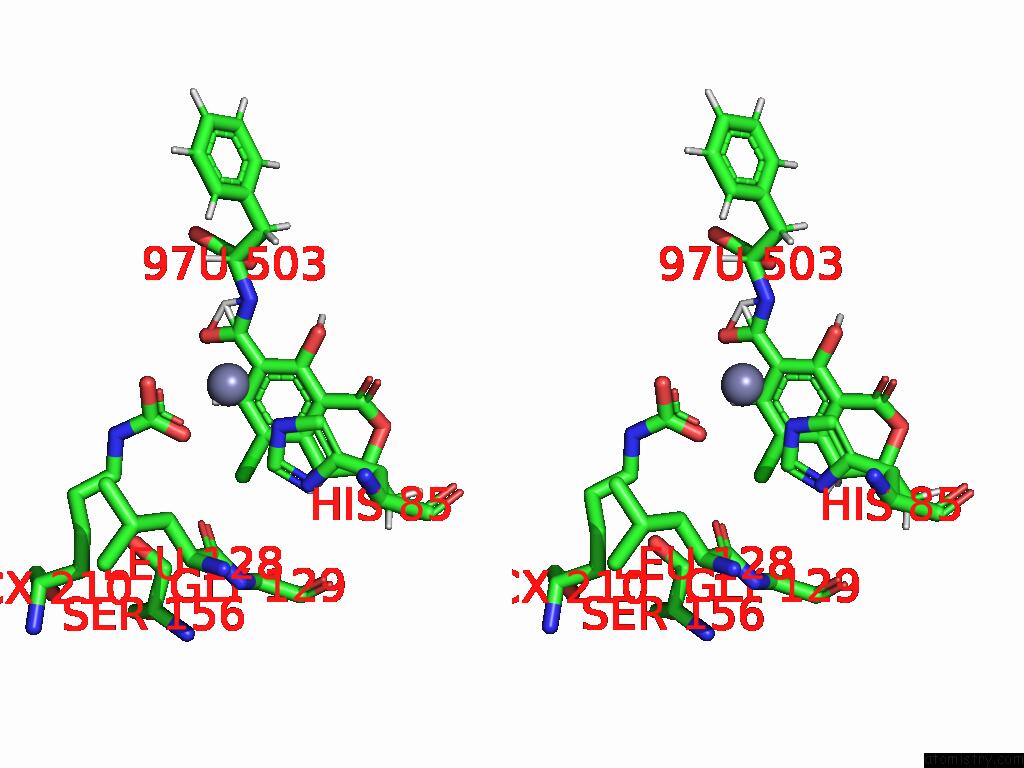
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase within 5.0Å range:
|
Chlorine binding site 5 out of 8 in 8yak
Go back to
Chlorine binding site 5 out
of 8 in the Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase
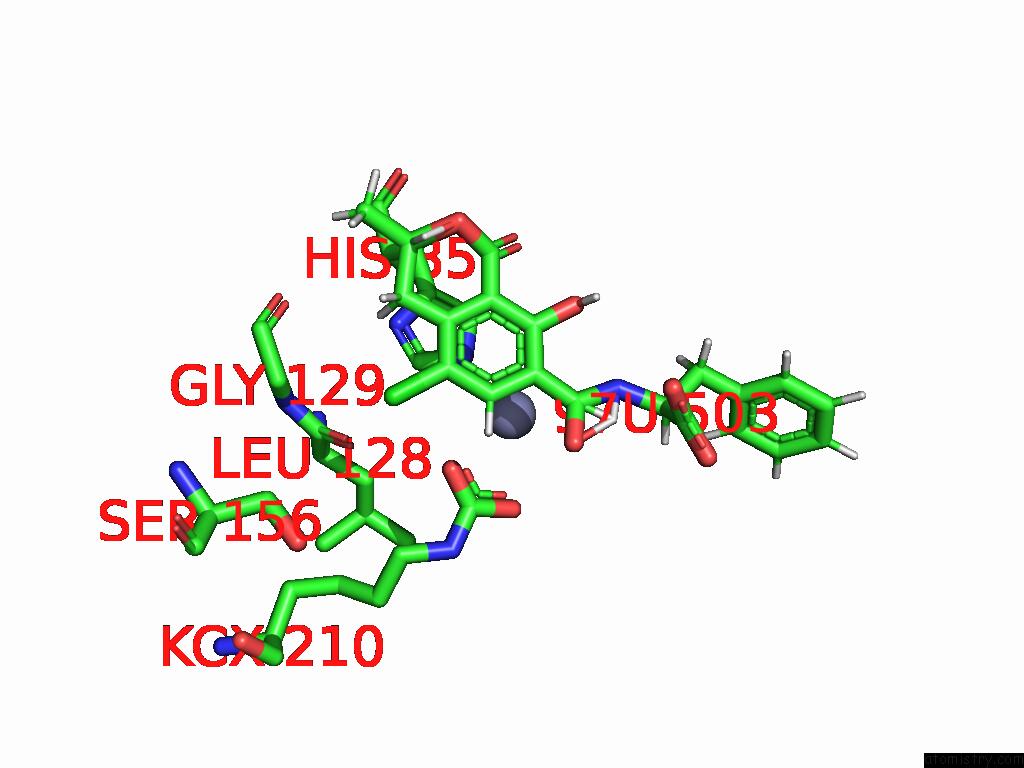
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase within 5.0Å range:
|
Chlorine binding site 6 out of 8 in 8yak
Go back to
Chlorine binding site 6 out
of 8 in the Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase
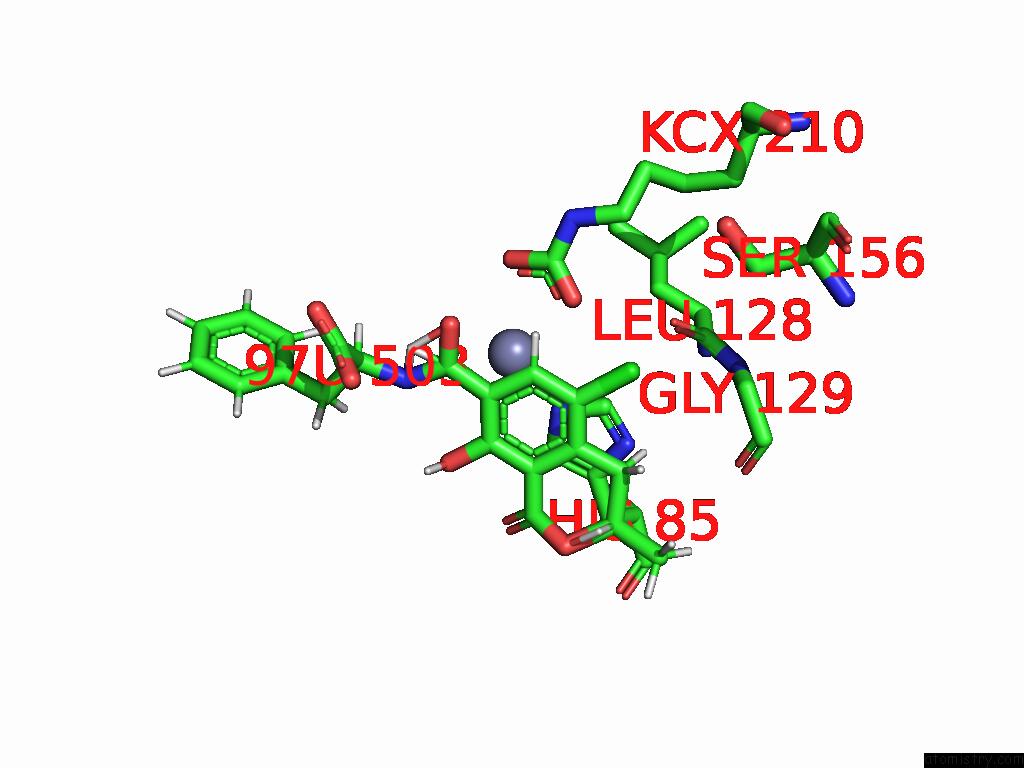
Mono view
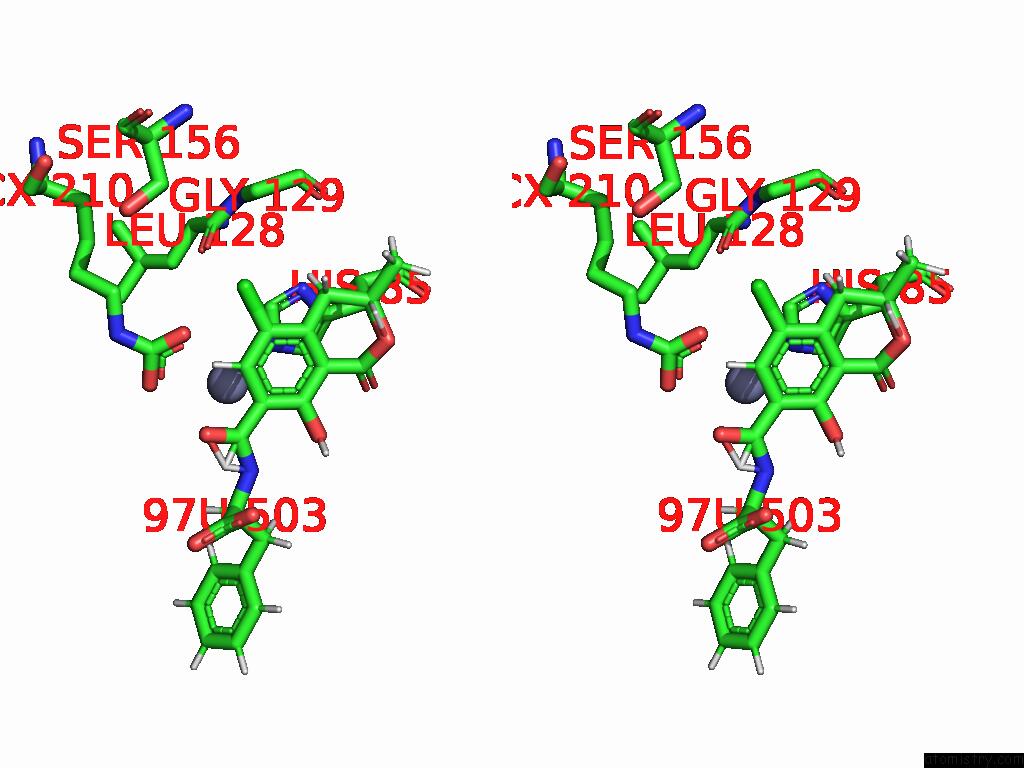
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 6 of Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase within 5.0Å range:
|
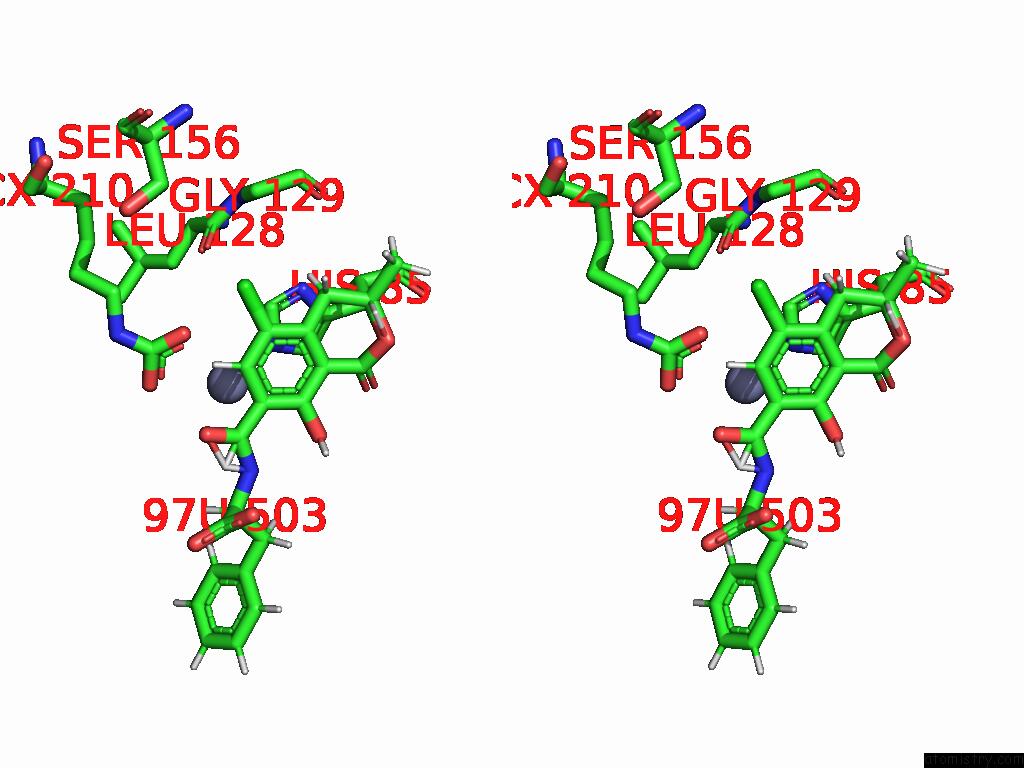
Chlorine binding site 7 out of 8 in 8yak
Go back to
Chlorine binding site 7 out
of 8 in the Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase
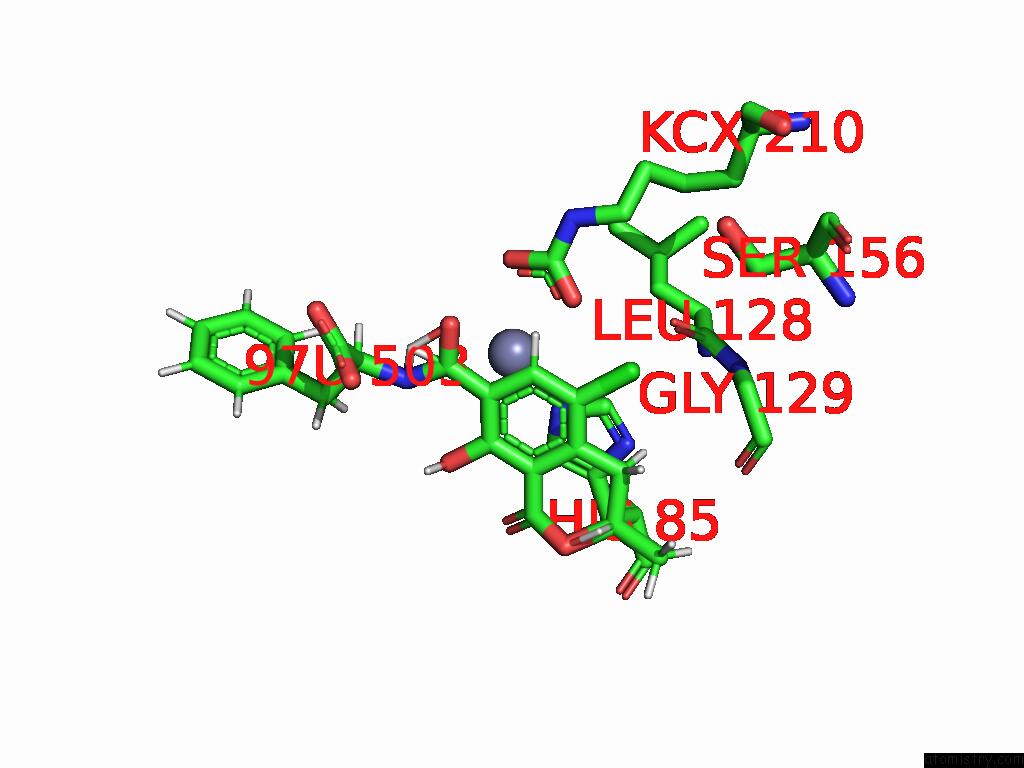
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 7 of Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase within 5.0Å range:
|
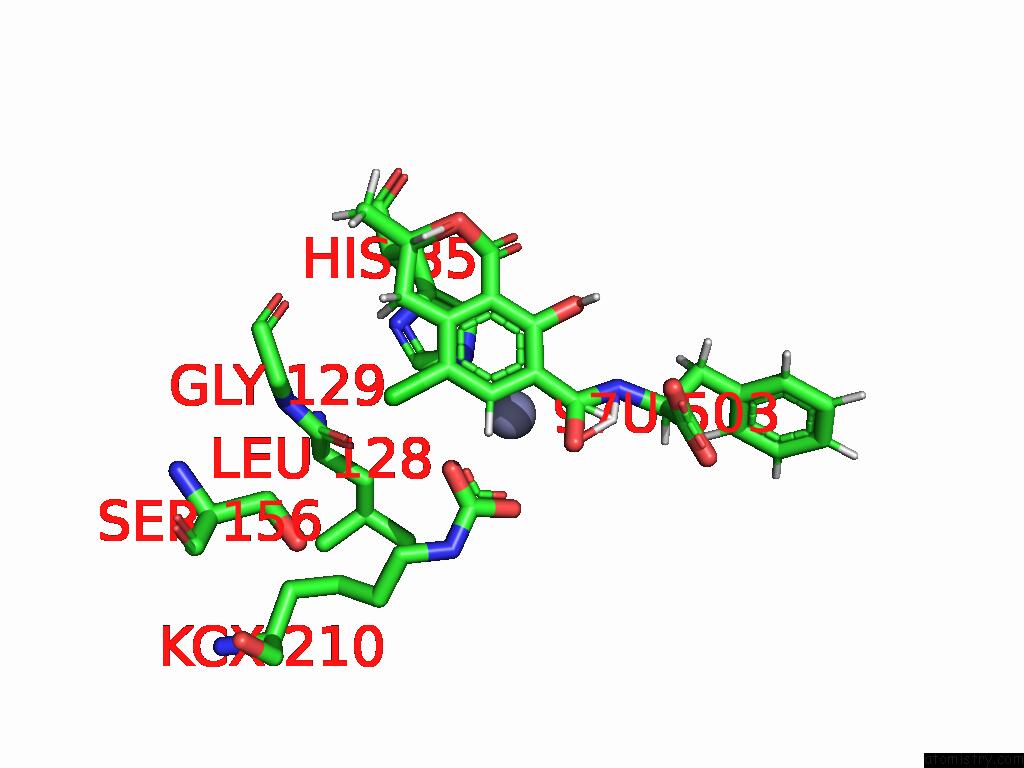
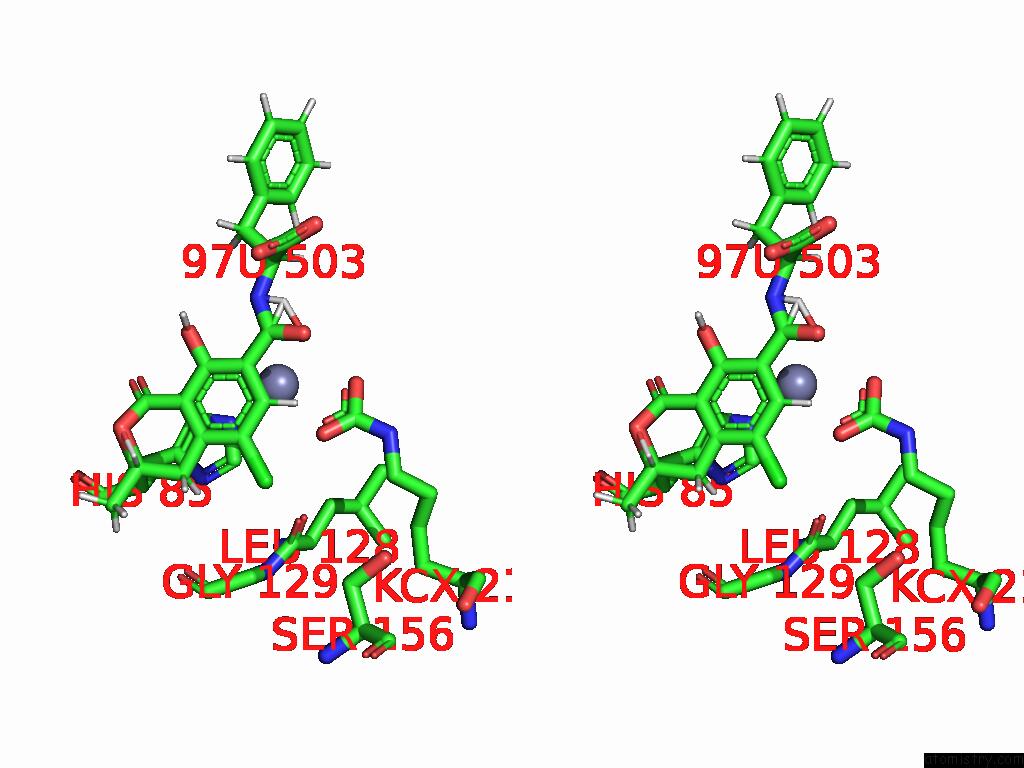
Chlorine binding site 8 out of 8 in 8yak
Go back to
Chlorine binding site 8 out
of 8 in the Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 8 of Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase within 5.0Å range:
|
Reference:
Y.Hu,
L.Dai,
Y.Xu,
D.Niu,
X.Yang,
Z.Xie,
P.Shen,
X.Li,
H.Li,
L.Zhang,
J.Min,
R.T.Guo,
C.C.Chen.
Functional Characterization and Structural Basis of An Efficient Ochratoxin A-Degrading Amidohydrolase. Int.J.Biol.Macromol. V. 278 34831 2024.
ISSN: ISSN 0141-8130
PubMed: 39163957
DOI: 10.1016/J.IJBIOMAC.2024.134831
Page generated: Sat Feb 8 17:23:52 2025
ISSN: ISSN 0141-8130
PubMed: 39163957
DOI: 10.1016/J.IJBIOMAC.2024.134831
Last articles
Zn in 9J0NZn in 9J0O
Zn in 9J0P
Zn in 9FJX
Zn in 9EKB
Zn in 9C0F
Zn in 9CAH
Zn in 9CH0
Zn in 9CH3
Zn in 9CH1