Chlorine »
PDB 1g0j-1gl7 »
1gac »
Chlorine in PDB 1gac: uc(Nmr) Structure of Asymmetric Homodimer of A82846B, A Glycopeptide Antibiotic, Complexed with Its Cell Wall Pentapeptide Fragment
Chlorine Binding Sites:
The binding sites of Chlorine atom in the uc(Nmr) Structure of Asymmetric Homodimer of A82846B, A Glycopeptide Antibiotic, Complexed with Its Cell Wall Pentapeptide Fragment
(pdb code 1gac). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 4 binding sites of Chlorine where determined in the uc(Nmr) Structure of Asymmetric Homodimer of A82846B, A Glycopeptide Antibiotic, Complexed with Its Cell Wall Pentapeptide Fragment, PDB code: 1gac:
Jump to Chlorine binding site number: 1; 2; 3; 4;
In total 4 binding sites of Chlorine where determined in the uc(Nmr) Structure of Asymmetric Homodimer of A82846B, A Glycopeptide Antibiotic, Complexed with Its Cell Wall Pentapeptide Fragment, PDB code: 1gac:
Jump to Chlorine binding site number: 1; 2; 3; 4;
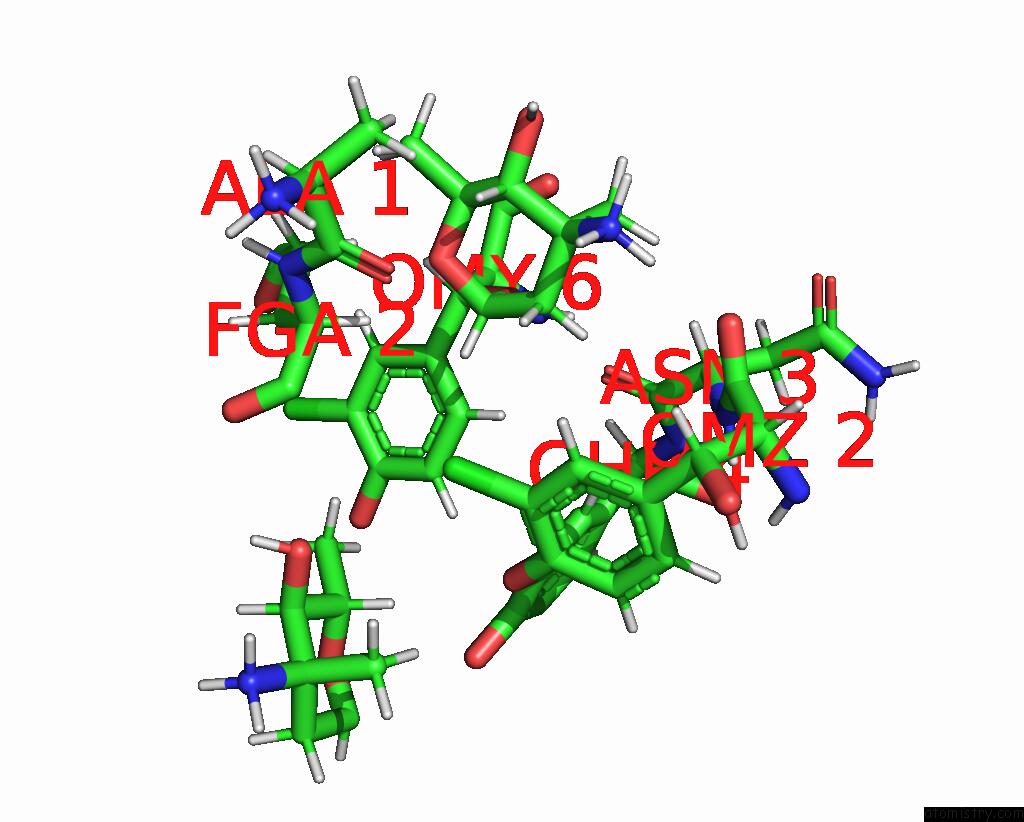
Chlorine binding site 1 out of 4 in 1gac
Go back to
Chlorine binding site 1 out
of 4 in the uc(Nmr) Structure of Asymmetric Homodimer of A82846B, A Glycopeptide Antibiotic, Complexed with Its Cell Wall Pentapeptide Fragment
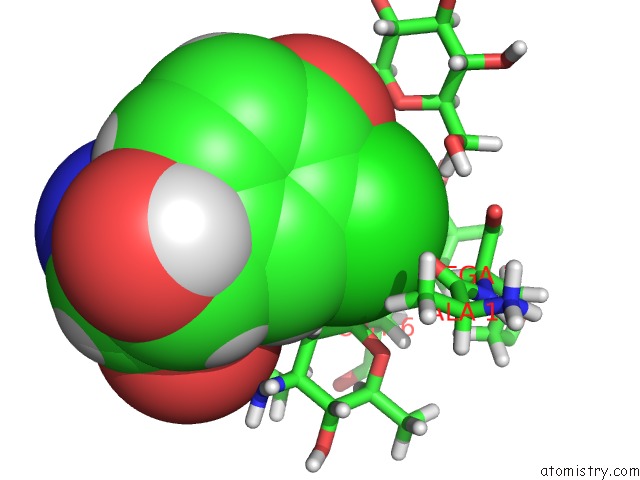
Mono view
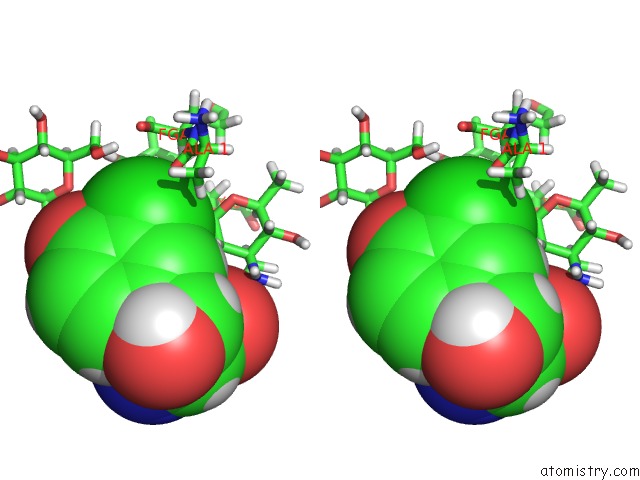
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of uc(Nmr) Structure of Asymmetric Homodimer of A82846B, A Glycopeptide Antibiotic, Complexed with Its Cell Wall Pentapeptide Fragment within 5.0Å range:
|
Chlorine binding site 2 out of 4 in 1gac
Go back to
Chlorine binding site 2 out
of 4 in the uc(Nmr) Structure of Asymmetric Homodimer of A82846B, A Glycopeptide Antibiotic, Complexed with Its Cell Wall Pentapeptide Fragment
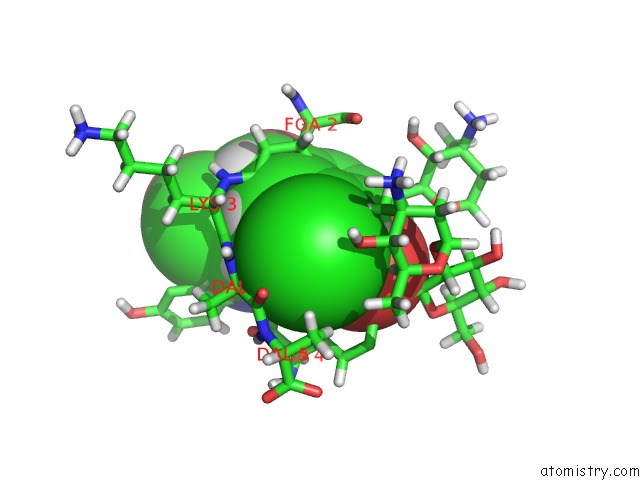
Mono view
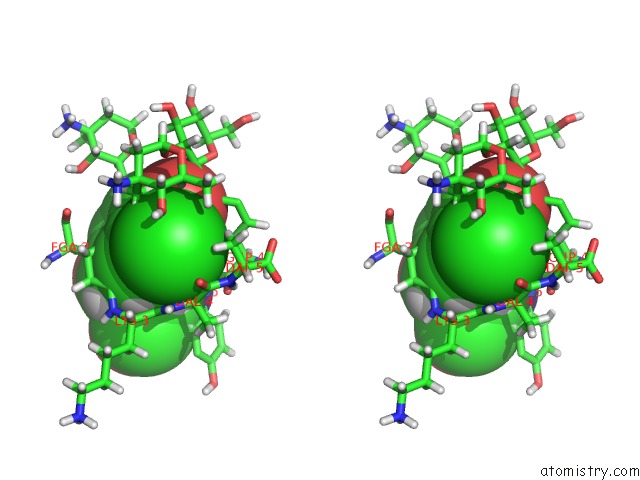
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of uc(Nmr) Structure of Asymmetric Homodimer of A82846B, A Glycopeptide Antibiotic, Complexed with Its Cell Wall Pentapeptide Fragment within 5.0Å range:
|
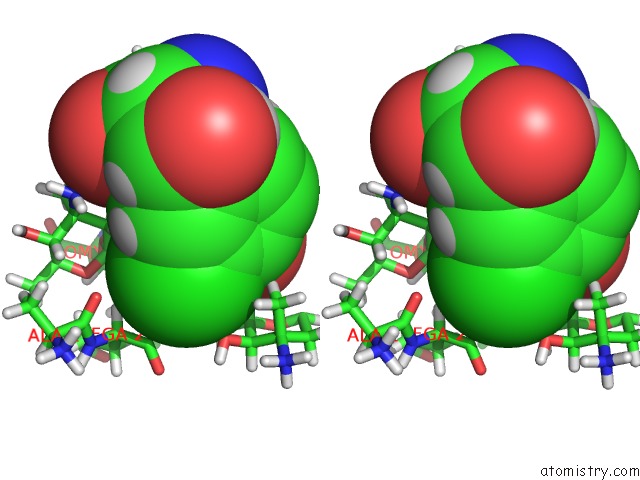
Chlorine binding site 3 out of 4 in 1gac
Go back to
Chlorine binding site 3 out
of 4 in the uc(Nmr) Structure of Asymmetric Homodimer of A82846B, A Glycopeptide Antibiotic, Complexed with Its Cell Wall Pentapeptide Fragment
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of uc(Nmr) Structure of Asymmetric Homodimer of A82846B, A Glycopeptide Antibiotic, Complexed with Its Cell Wall Pentapeptide Fragment within 5.0Å range:
|
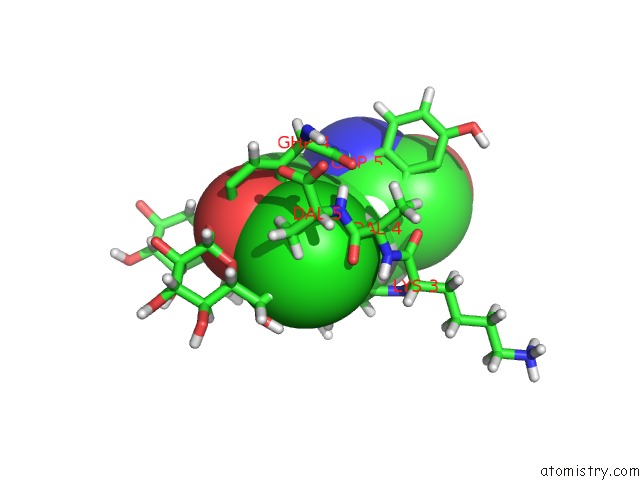
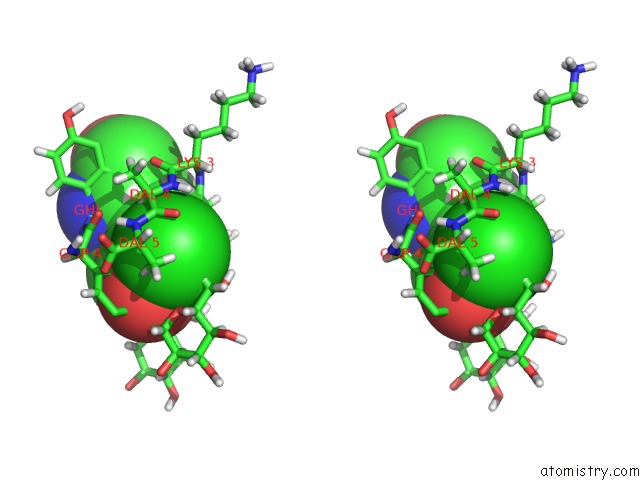
Chlorine binding site 4 out of 4 in 1gac
Go back to
Chlorine binding site 4 out
of 4 in the uc(Nmr) Structure of Asymmetric Homodimer of A82846B, A Glycopeptide Antibiotic, Complexed with Its Cell Wall Pentapeptide Fragment
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of uc(Nmr) Structure of Asymmetric Homodimer of A82846B, A Glycopeptide Antibiotic, Complexed with Its Cell Wall Pentapeptide Fragment within 5.0Å range:
|
Reference:
W.G.Prowse,
A.D.Kline,
M.A.Skelton,
R.J.Loncharich.
Conformation of A82846B, A Glycopeptide Antibiotic, Complexed with Its Cell Wall Fragment: An Asymmetric Homodimer Determined Using uc(Nmr) Spectroscopy. Biochemistry V. 34 9632 1995.
ISSN: ISSN 0006-2960
PubMed: 7626632
DOI: 10.1021/BI00029A041
Page generated: Fri Jul 19 22:09:55 2024
ISSN: ISSN 0006-2960
PubMed: 7626632
DOI: 10.1021/BI00029A041
Last articles
Zn in 9J0NZn in 9J0O
Zn in 9J0P
Zn in 9FJX
Zn in 9EKB
Zn in 9C0F
Zn in 9CAH
Zn in 9CH0
Zn in 9CH3
Zn in 9CH1