Chlorine »
PDB 1i3l-1iqi »
1ihu »
Chlorine in PDB 1ihu: Crystal Structure of the Escherichia Coli Arsenite-Translocating Atpase in Complex with Mg-Adp-ALF3
Enzymatic activity of Crystal Structure of the Escherichia Coli Arsenite-Translocating Atpase in Complex with Mg-Adp-ALF3
All present enzymatic activity of Crystal Structure of the Escherichia Coli Arsenite-Translocating Atpase in Complex with Mg-Adp-ALF3:
3.6.3.16;
3.6.3.16;
Protein crystallography data
The structure of Crystal Structure of the Escherichia Coli Arsenite-Translocating Atpase in Complex with Mg-Adp-ALF3, PDB code: 1ihu
was solved by
T.Zhou,
S.Radaev,
B.P.Rosen,
D.L.Gatti,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 25.15 / 2.15 |
| Space group | I 2 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 73.897, 75.945, 222.607, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 21.5 / 26.2 |
Other elements in 1ihu:
The structure of Crystal Structure of the Escherichia Coli Arsenite-Translocating Atpase in Complex with Mg-Adp-ALF3 also contains other interesting chemical elements:
| Fluorine | (F) | 3 atoms |
| Magnesium | (Mg) | 2 atoms |
| Aluminium | (Al) | 1 atom |
| Cadmium | (Cd) | 8 atoms |
| Arsenic | (As) | 1 atom |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of the Escherichia Coli Arsenite-Translocating Atpase in Complex with Mg-Adp-ALF3
(pdb code 1ihu). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 3 binding sites of Chlorine where determined in the Crystal Structure of the Escherichia Coli Arsenite-Translocating Atpase in Complex with Mg-Adp-ALF3, PDB code: 1ihu:
Jump to Chlorine binding site number: 1; 2; 3;
In total 3 binding sites of Chlorine where determined in the Crystal Structure of the Escherichia Coli Arsenite-Translocating Atpase in Complex with Mg-Adp-ALF3, PDB code: 1ihu:
Jump to Chlorine binding site number: 1; 2; 3;
Chlorine binding site 1 out of 3 in 1ihu
Go back to
Chlorine binding site 1 out
of 3 in the Crystal Structure of the Escherichia Coli Arsenite-Translocating Atpase in Complex with Mg-Adp-ALF3
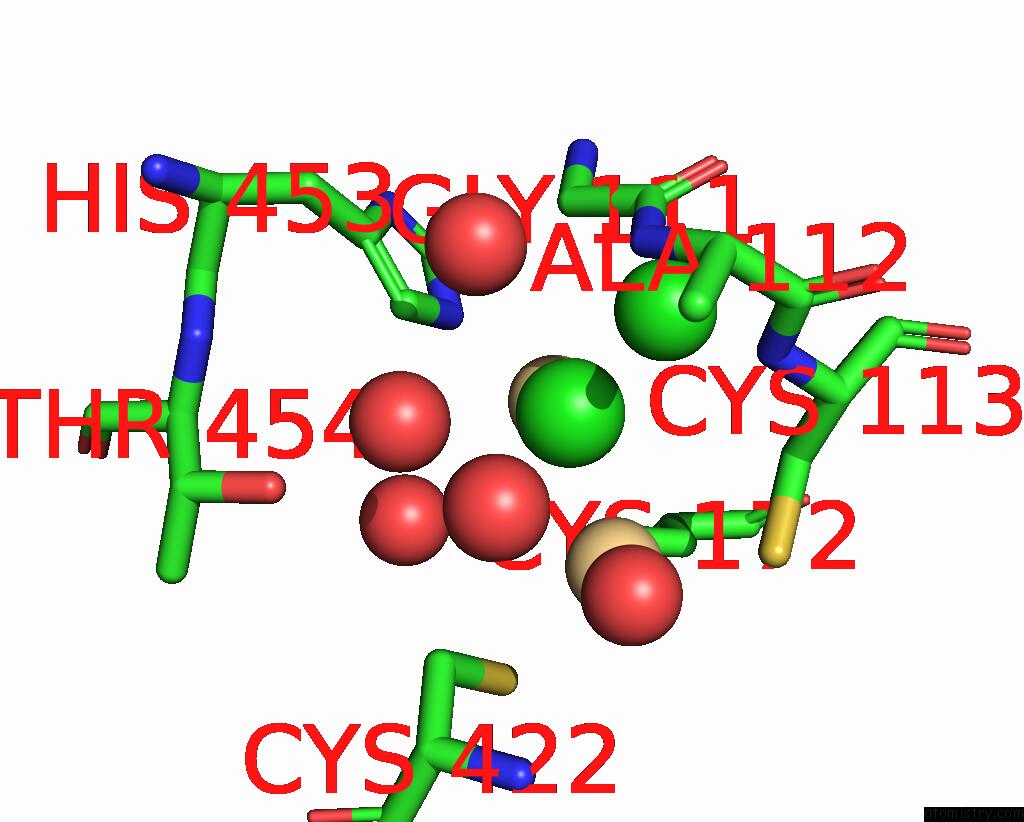
Mono view
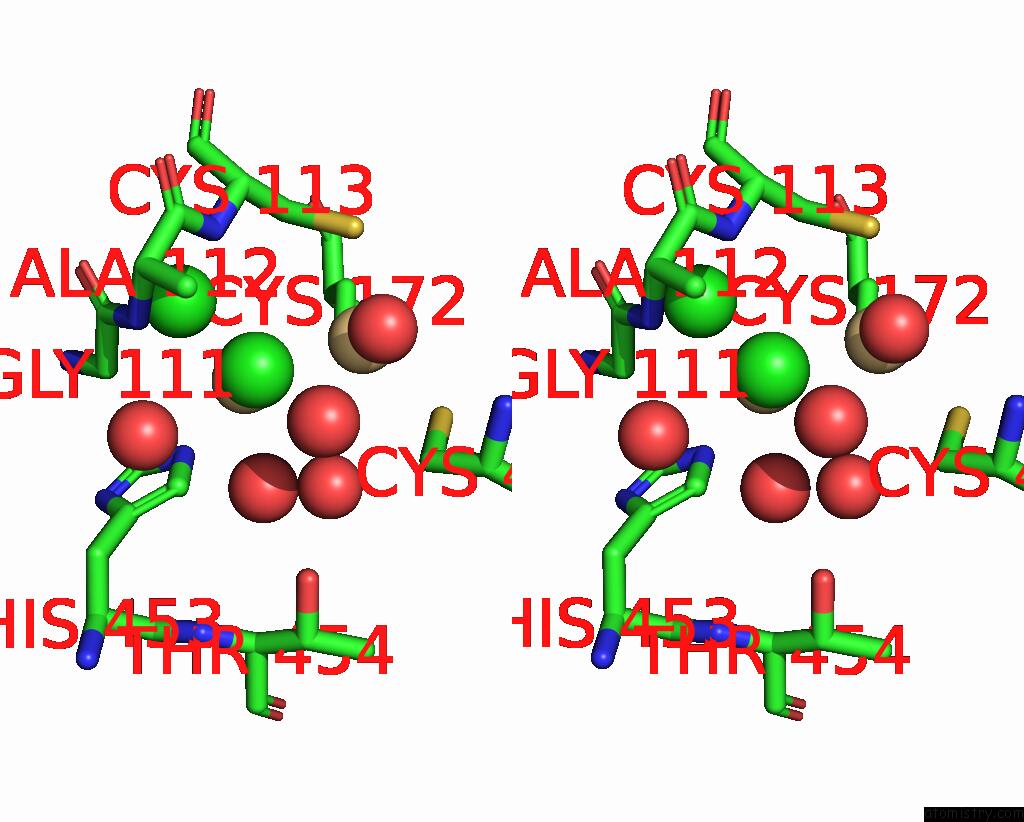
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of the Escherichia Coli Arsenite-Translocating Atpase in Complex with Mg-Adp-ALF3 within 5.0Å range:
|
Chlorine binding site 2 out of 3 in 1ihu
Go back to
Chlorine binding site 2 out
of 3 in the Crystal Structure of the Escherichia Coli Arsenite-Translocating Atpase in Complex with Mg-Adp-ALF3
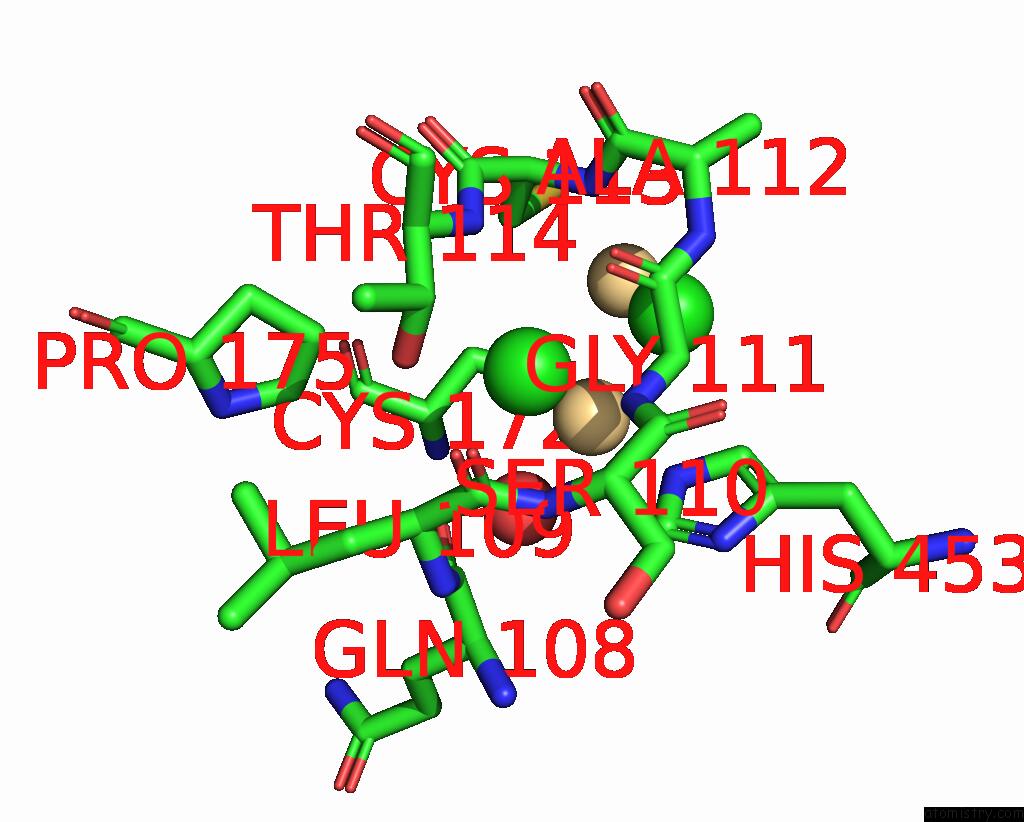
Mono view
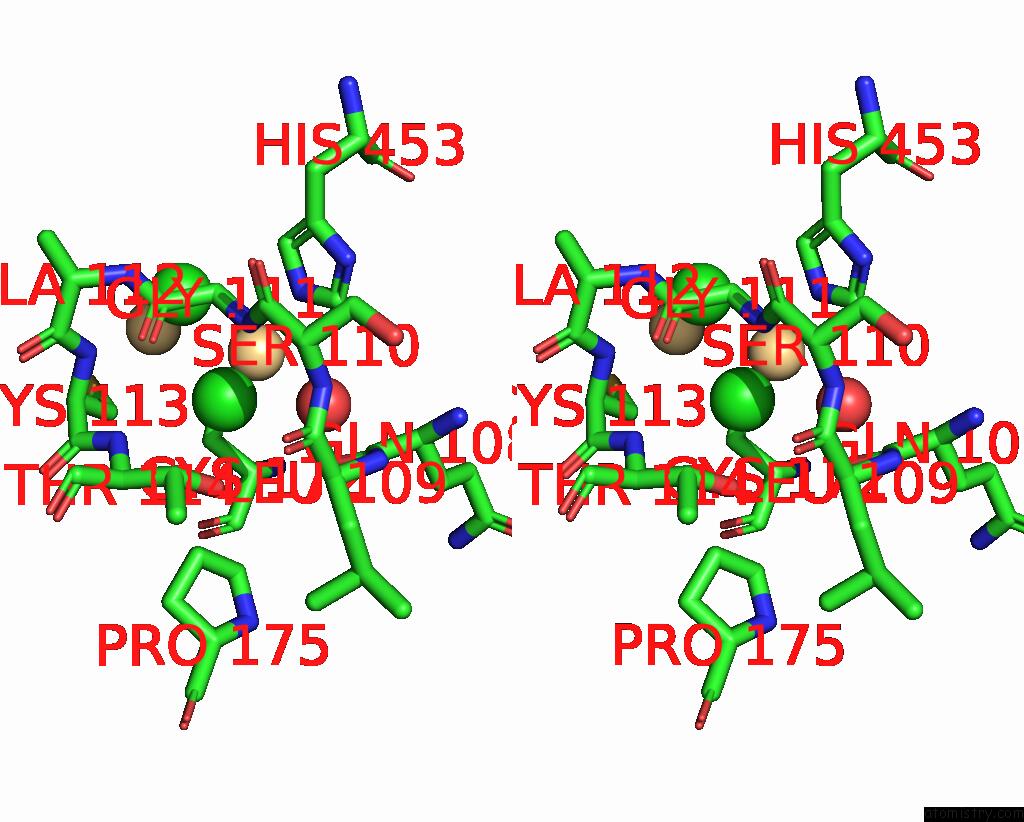
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of the Escherichia Coli Arsenite-Translocating Atpase in Complex with Mg-Adp-ALF3 within 5.0Å range:
|
Chlorine binding site 3 out of 3 in 1ihu
Go back to
Chlorine binding site 3 out
of 3 in the Crystal Structure of the Escherichia Coli Arsenite-Translocating Atpase in Complex with Mg-Adp-ALF3
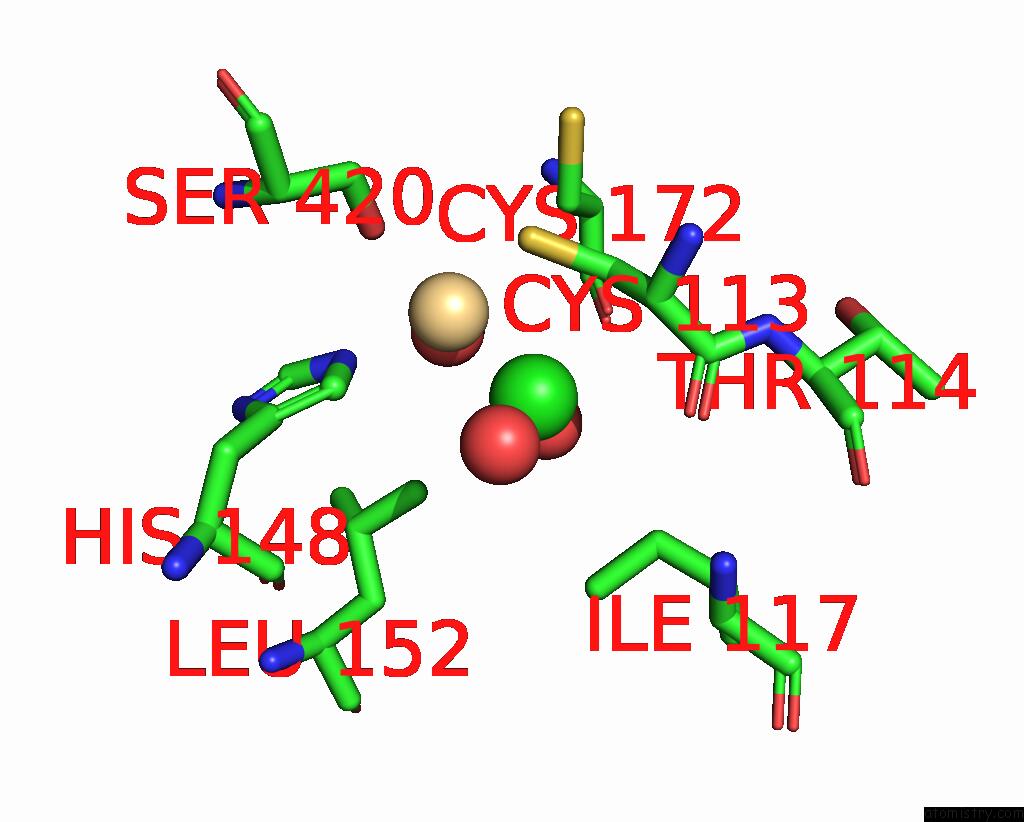
Mono view
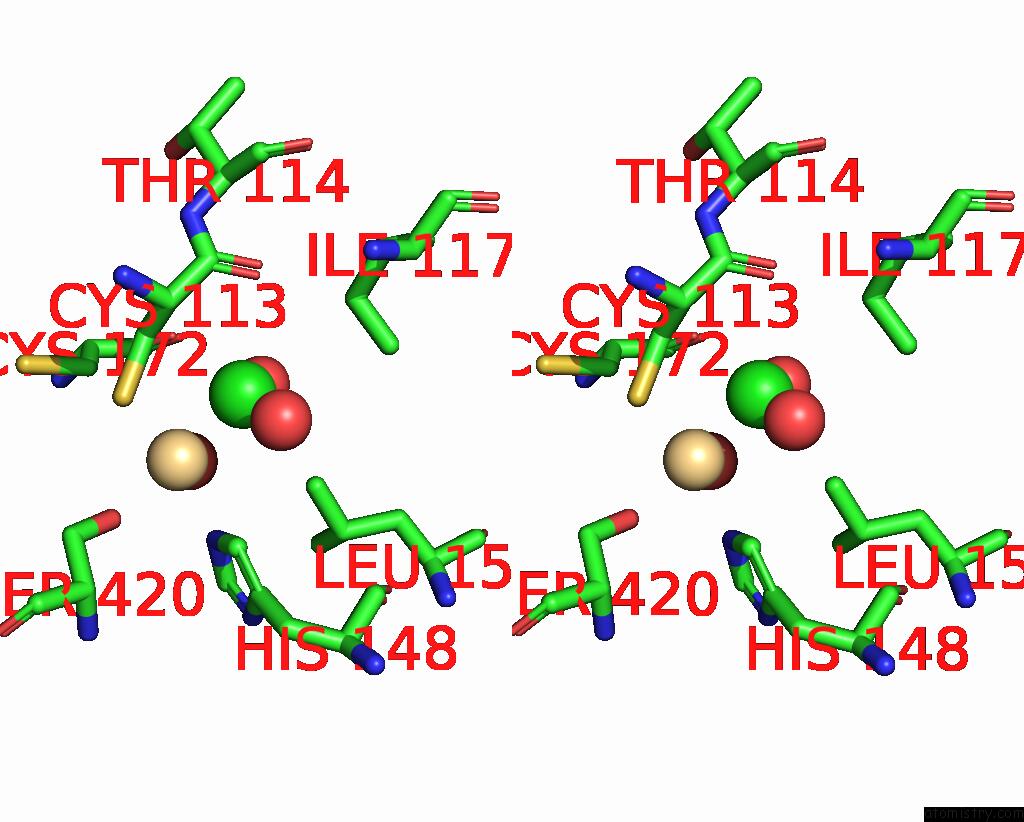
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Crystal Structure of the Escherichia Coli Arsenite-Translocating Atpase in Complex with Mg-Adp-ALF3 within 5.0Å range:
|
Reference:
T.Zhou,
S.Radaev,
B.P.Rosen,
D.L.Gatti.
Conformational Changes in Four Regions of the Escherichia Coli Arsa Atpase Link Atp Hydrolysis to Ion Translocation. J.Biol.Chem. V. 276 30414 2001.
ISSN: ISSN 0021-9258
PubMed: 11395509
DOI: 10.1074/JBC.M103671200
Page generated: Thu Jul 10 17:23:51 2025
ISSN: ISSN 0021-9258
PubMed: 11395509
DOI: 10.1074/JBC.M103671200
Last articles
F in 4JLNF in 4JLT
F in 4JLM
F in 4JLG
F in 4JJU
F in 4JLK
F in 4JKV
F in 4JLJ
F in 4JJS
F in 4JII