Chlorine »
PDB 1wbq-1wvf »
1wlu »
Chlorine in PDB 1wlu: Crystal Structure of TT0310 Protein From Thermus Thermophilus HB8
Protein crystallography data
The structure of Crystal Structure of TT0310 Protein From Thermus Thermophilus HB8, PDB code: 1wlu
was solved by
N.Kunishima,
M.Sugahara,
M.Miyano,
Riken Structural Genomics/Proteomicsinitiative (Rsgi),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 25.00 / 1.45 |
| Space group | I 41 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 76.804, 76.804, 155.701, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 18.8 / 19.4 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of TT0310 Protein From Thermus Thermophilus HB8
(pdb code 1wlu). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the Crystal Structure of TT0310 Protein From Thermus Thermophilus HB8, PDB code: 1wlu:
In total only one binding site of Chlorine was determined in the Crystal Structure of TT0310 Protein From Thermus Thermophilus HB8, PDB code: 1wlu:
Chlorine binding site 1 out of 1 in 1wlu
Go back to
Chlorine binding site 1 out
of 1 in the Crystal Structure of TT0310 Protein From Thermus Thermophilus HB8
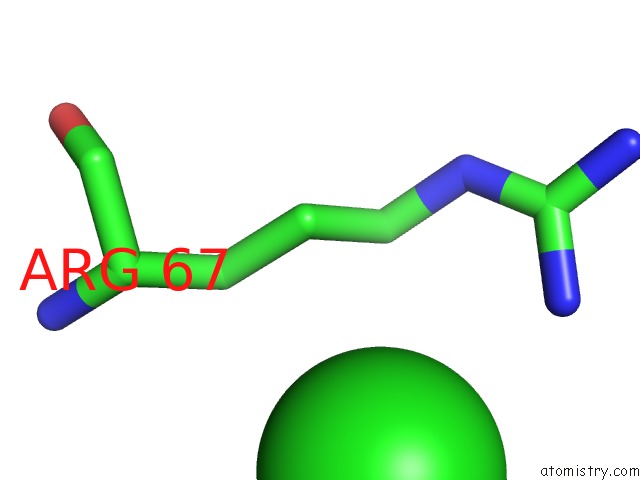
Mono view
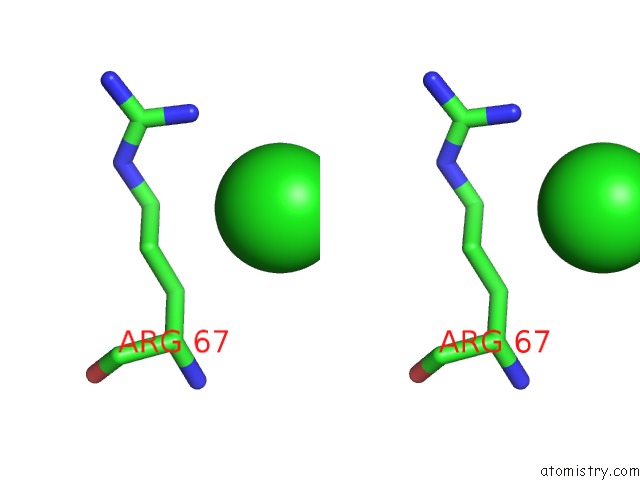
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of TT0310 Protein From Thermus Thermophilus HB8 within 5.0Å range:
|
Reference:
N.Kunishima,
Y.Asada,
M.Sugahara,
J.Ishijima,
Y.Nodake,
M.Sugahara,
M.Miyano,
S.Kuramitsu,
S.Yokoyama,
M.Sugahara.
A Novel Induced-Fit Reaction Mechanism of Asymmetric Hot Dog Thioesterase Paai J.Mol.Biol. V. 352 212 2005.
ISSN: ISSN 0022-2836
PubMed: 16061252
DOI: 10.1016/J.JMB.2005.07.008
Page generated: Thu Jul 10 20:17:42 2025
ISSN: ISSN 0022-2836
PubMed: 16061252
DOI: 10.1016/J.JMB.2005.07.008
Last articles
Cl in 5UU4Cl in 5URS
Cl in 5UU3
Cl in 5URQ
Cl in 5UTN
Cl in 5USS
Cl in 5URU
Cl in 5USQ
Cl in 5UQH
Cl in 5UQG