Chlorine »
PDB 2etd-2f81 »
2f2q »
Chlorine in PDB 2f2q: High Resolution Crystal Strcuture of T4 Lysozyme Mutant L20R63/A Liganded to Guanidinium Ion
Enzymatic activity of High Resolution Crystal Strcuture of T4 Lysozyme Mutant L20R63/A Liganded to Guanidinium Ion
All present enzymatic activity of High Resolution Crystal Strcuture of T4 Lysozyme Mutant L20R63/A Liganded to Guanidinium Ion:
3.2.1.17;
3.2.1.17;
Protein crystallography data
The structure of High Resolution Crystal Strcuture of T4 Lysozyme Mutant L20R63/A Liganded to Guanidinium Ion, PDB code: 2f2q
was solved by
M.S.Yousef,
N.Bischoff,
C.M.Dyer,
W.A.Baase,
B.W.Matthews,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 30.00 / 1.45 |
| Space group | P 32 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 60.712, 60.712, 95.352, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 20 / 22.3 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the High Resolution Crystal Strcuture of T4 Lysozyme Mutant L20R63/A Liganded to Guanidinium Ion
(pdb code 2f2q). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the High Resolution Crystal Strcuture of T4 Lysozyme Mutant L20R63/A Liganded to Guanidinium Ion, PDB code: 2f2q:
In total only one binding site of Chlorine was determined in the High Resolution Crystal Strcuture of T4 Lysozyme Mutant L20R63/A Liganded to Guanidinium Ion, PDB code: 2f2q:
Chlorine binding site 1 out of 1 in 2f2q
Go back to
Chlorine binding site 1 out
of 1 in the High Resolution Crystal Strcuture of T4 Lysozyme Mutant L20R63/A Liganded to Guanidinium Ion
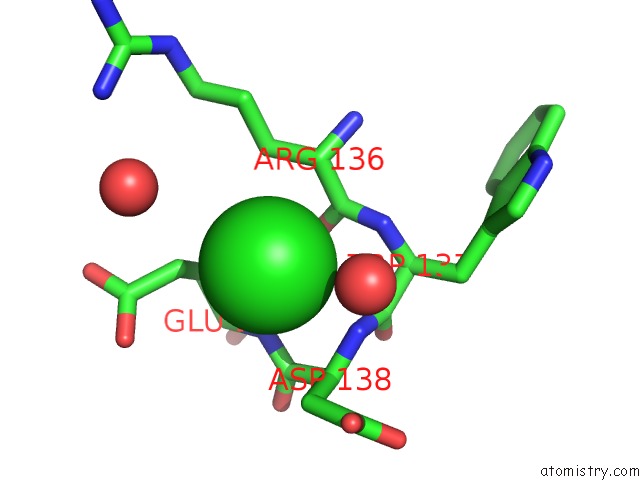
Mono view
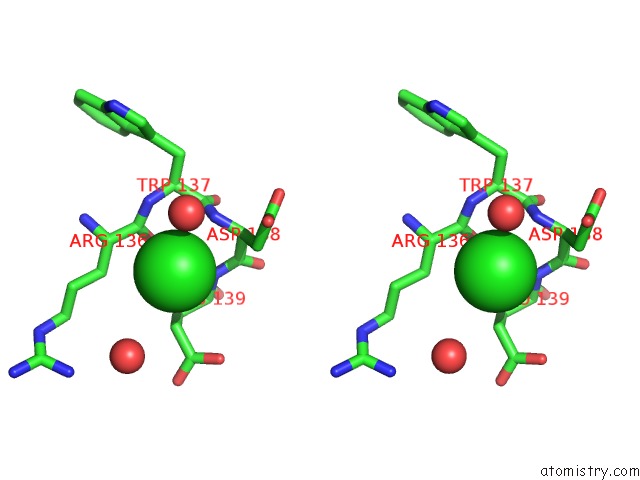
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of High Resolution Crystal Strcuture of T4 Lysozyme Mutant L20R63/A Liganded to Guanidinium Ion within 5.0Å range:
|
Reference:
M.S.Yousef,
N.Bischoff,
C.M.Dyer,
W.A.Baase,
B.W.Matthews.
Guanidinium Derivatives Bind Preferentially and Trigger Long-Distance Conformational Changes in An Engineered T4 Lysozyme. Protein Sci. V. 15 853 2006.
ISSN: ISSN 0961-8368
PubMed: 16600969
DOI: 10.1110/PS.052020606
Page generated: Thu Jul 10 22:02:26 2025
ISSN: ISSN 0961-8368
PubMed: 16600969
DOI: 10.1110/PS.052020606
Last articles
Cl in 5SIOCl in 5SID
Cl in 5SIE
Cl in 5SIL
Cl in 5SHT
Cl in 5SIF
Cl in 5SHY
Cl in 5SHL
Cl in 5SHM
Cl in 5SHG