Chlorine »
PDB 2kug-2nwh »
2nv2 »
Chlorine in PDB 2nv2: Structure of the Plp Synthase Complex PDX1/2 (Yaad/E) From Bacillus Subtilis
Protein crystallography data
The structure of Structure of the Plp Synthase Complex PDX1/2 (Yaad/E) From Bacillus Subtilis, PDB code: 2nv2
was solved by
M.Strohmeier,
I.Tews,
I.Sinning,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 50.00 / 2.12 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 93.507, 259.009, 144.962, 90.00, 92.13, 90.00 |
| R / Rfree (%) | 14.6 / 19.8 |
Chlorine Binding Sites:
Pages:
>>> Page 1 <<< Page 2, Binding sites: 11 - 12;Binding sites:
The binding sites of Chlorine atom in the Structure of the Plp Synthase Complex PDX1/2 (Yaad/E) From Bacillus Subtilis (pdb code 2nv2). This binding sites where shown within 5.0 Angstroms radius around Chlorine atom.In total 12 binding sites of Chlorine where determined in the Structure of the Plp Synthase Complex PDX1/2 (Yaad/E) From Bacillus Subtilis, PDB code: 2nv2:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9; 10;
Chlorine binding site 1 out of 12 in 2nv2
Go back to
Chlorine binding site 1 out
of 12 in the Structure of the Plp Synthase Complex PDX1/2 (Yaad/E) From Bacillus Subtilis
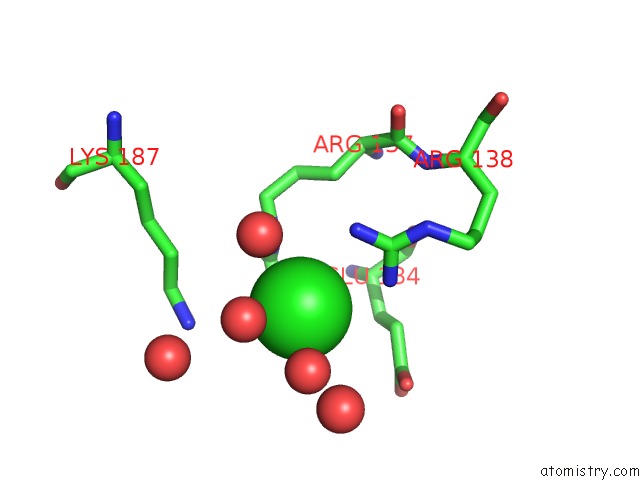
Mono view
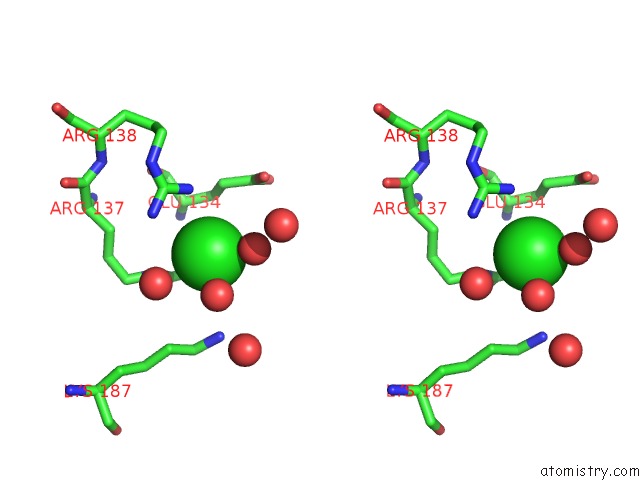
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Structure of the Plp Synthase Complex PDX1/2 (Yaad/E) From Bacillus Subtilis within 5.0Å range:
|
Chlorine binding site 2 out of 12 in 2nv2
Go back to
Chlorine binding site 2 out
of 12 in the Structure of the Plp Synthase Complex PDX1/2 (Yaad/E) From Bacillus Subtilis
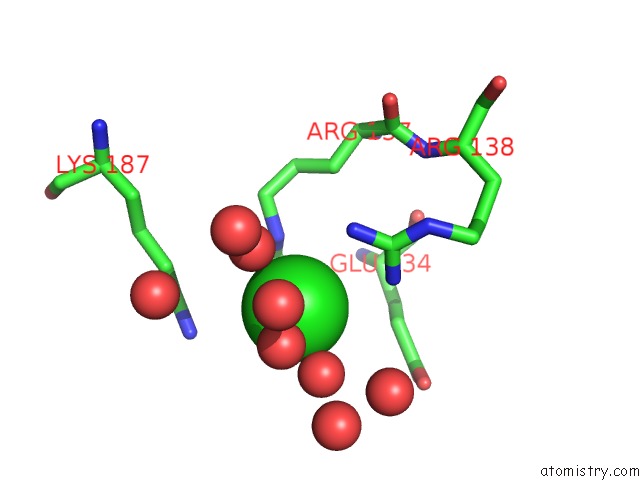
Mono view
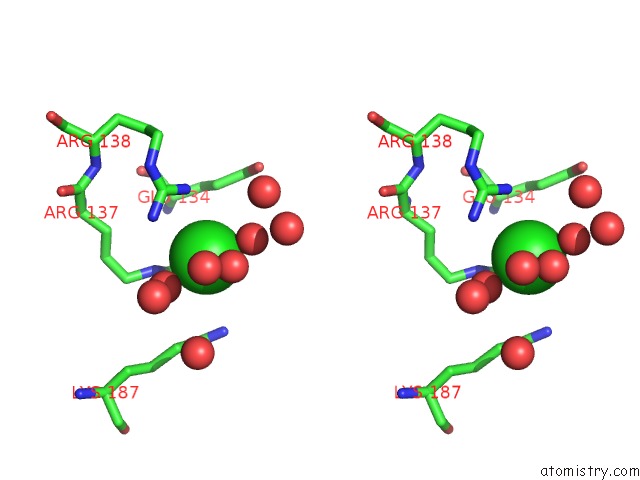
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Structure of the Plp Synthase Complex PDX1/2 (Yaad/E) From Bacillus Subtilis within 5.0Å range:
|
Chlorine binding site 3 out of 12 in 2nv2
Go back to
Chlorine binding site 3 out
of 12 in the Structure of the Plp Synthase Complex PDX1/2 (Yaad/E) From Bacillus Subtilis
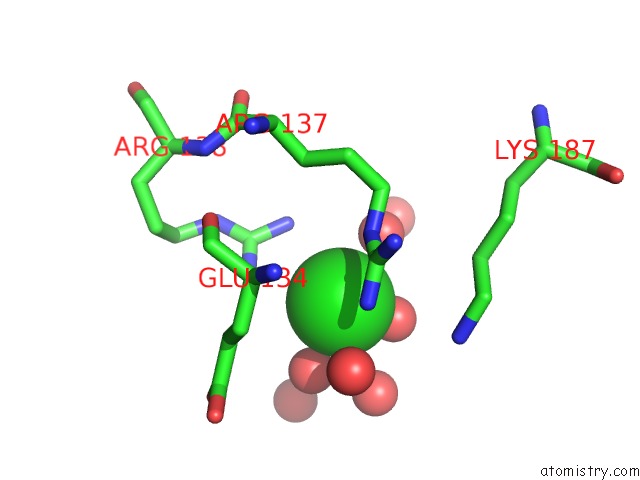
Mono view
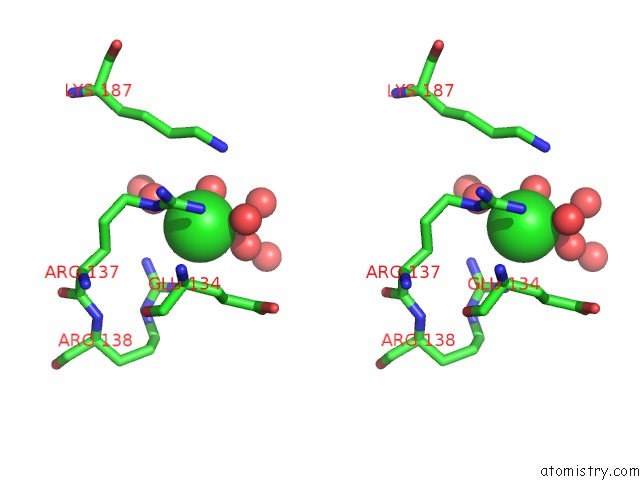
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Structure of the Plp Synthase Complex PDX1/2 (Yaad/E) From Bacillus Subtilis within 5.0Å range:
|
Chlorine binding site 4 out of 12 in 2nv2
Go back to
Chlorine binding site 4 out
of 12 in the Structure of the Plp Synthase Complex PDX1/2 (Yaad/E) From Bacillus Subtilis
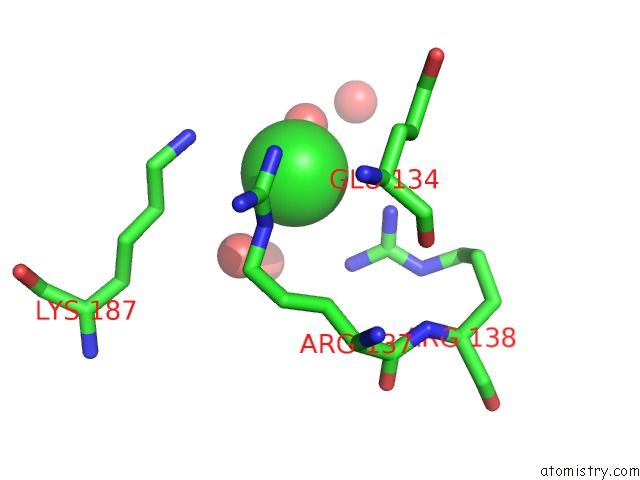
Mono view
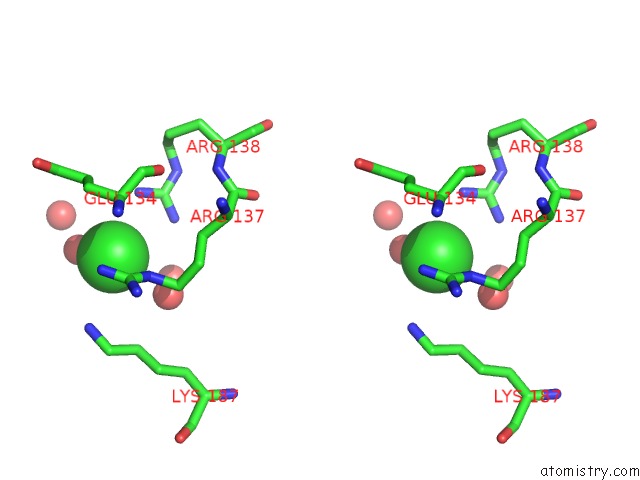
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Structure of the Plp Synthase Complex PDX1/2 (Yaad/E) From Bacillus Subtilis within 5.0Å range:
|
Chlorine binding site 5 out of 12 in 2nv2
Go back to
Chlorine binding site 5 out
of 12 in the Structure of the Plp Synthase Complex PDX1/2 (Yaad/E) From Bacillus Subtilis
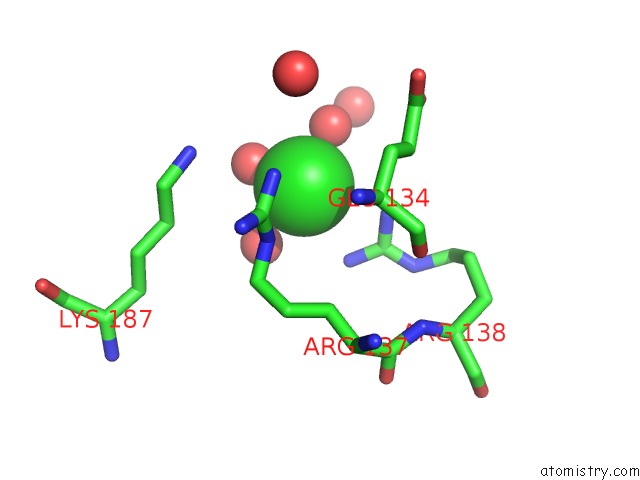
Mono view
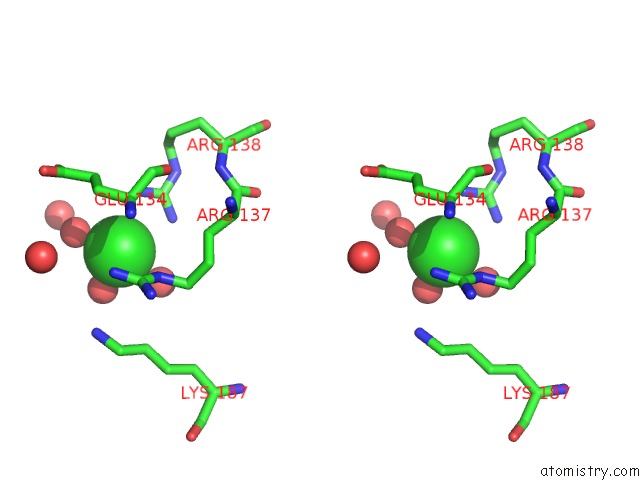
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Structure of the Plp Synthase Complex PDX1/2 (Yaad/E) From Bacillus Subtilis within 5.0Å range:
|
Chlorine binding site 6 out of 12 in 2nv2
Go back to
Chlorine binding site 6 out
of 12 in the Structure of the Plp Synthase Complex PDX1/2 (Yaad/E) From Bacillus Subtilis
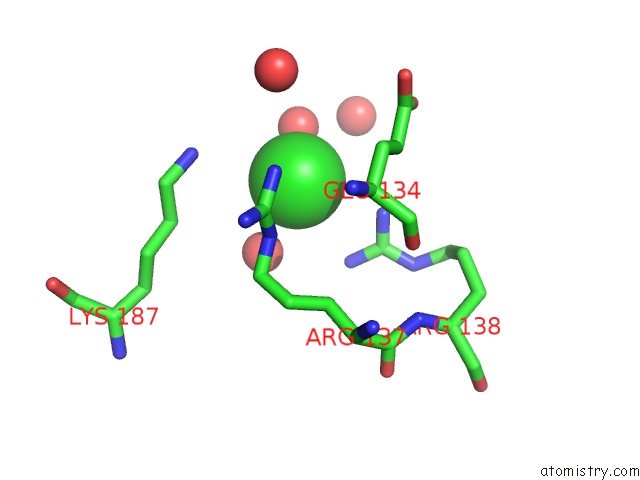
Mono view
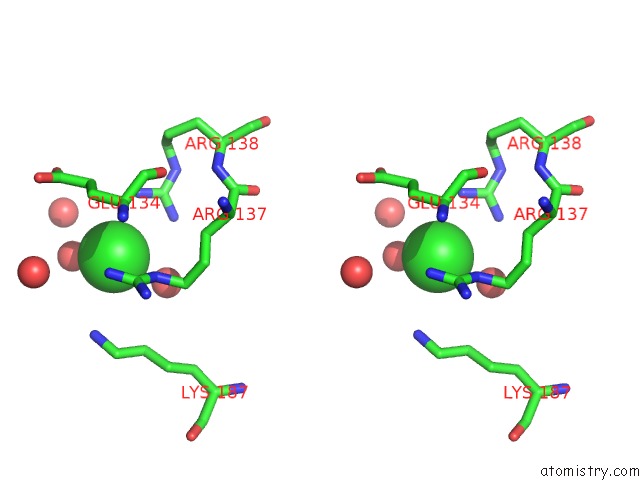
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 6 of Structure of the Plp Synthase Complex PDX1/2 (Yaad/E) From Bacillus Subtilis within 5.0Å range:
|
Chlorine binding site 7 out of 12 in 2nv2
Go back to
Chlorine binding site 7 out
of 12 in the Structure of the Plp Synthase Complex PDX1/2 (Yaad/E) From Bacillus Subtilis
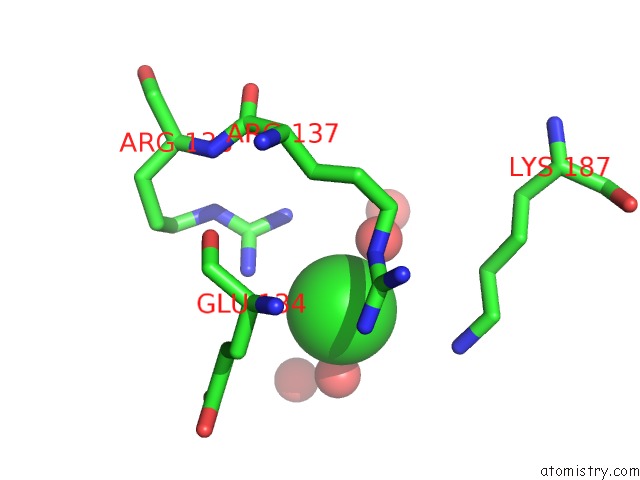
Mono view
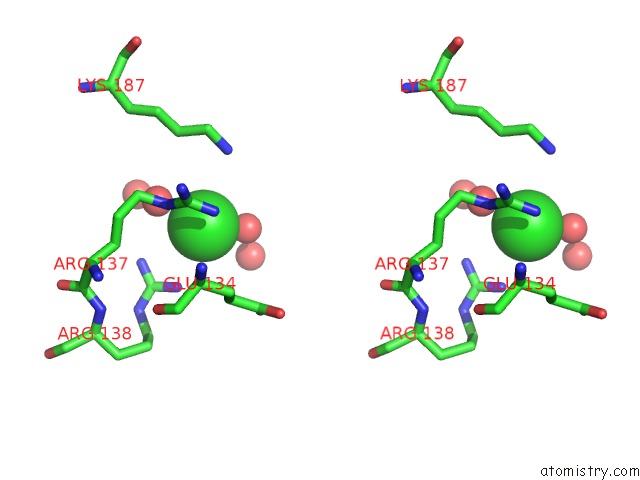
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 7 of Structure of the Plp Synthase Complex PDX1/2 (Yaad/E) From Bacillus Subtilis within 5.0Å range:
|
Chlorine binding site 8 out of 12 in 2nv2
Go back to
Chlorine binding site 8 out
of 12 in the Structure of the Plp Synthase Complex PDX1/2 (Yaad/E) From Bacillus Subtilis
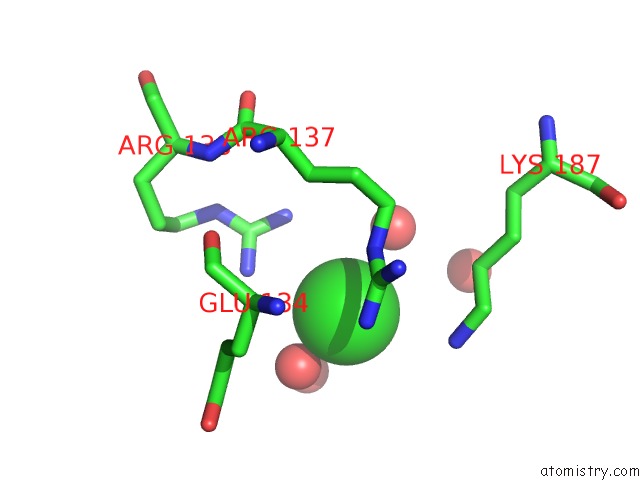
Mono view
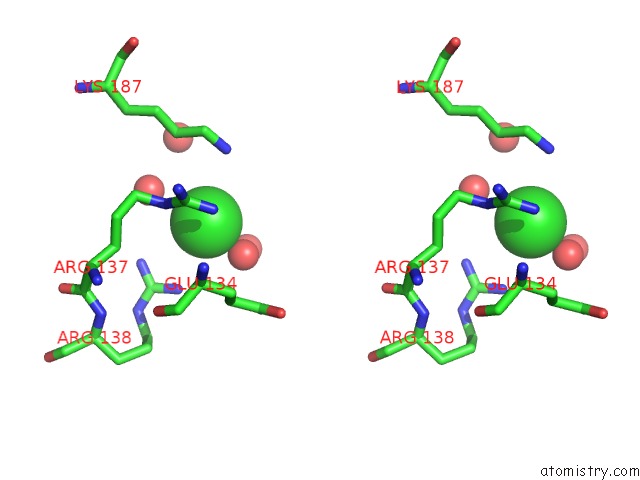
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 8 of Structure of the Plp Synthase Complex PDX1/2 (Yaad/E) From Bacillus Subtilis within 5.0Å range:
|
Chlorine binding site 9 out of 12 in 2nv2
Go back to
Chlorine binding site 9 out
of 12 in the Structure of the Plp Synthase Complex PDX1/2 (Yaad/E) From Bacillus Subtilis
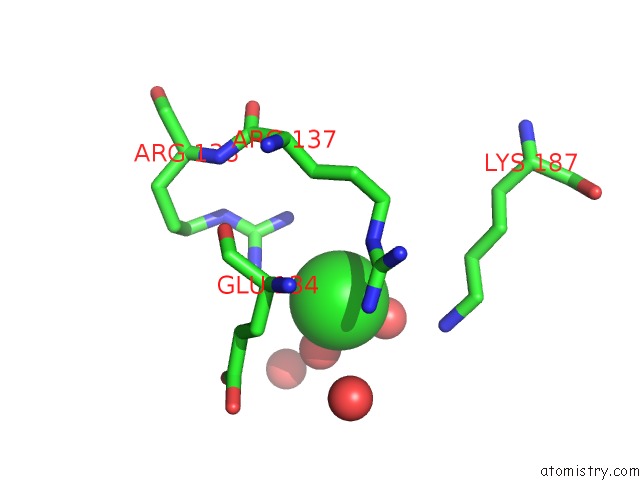
Mono view
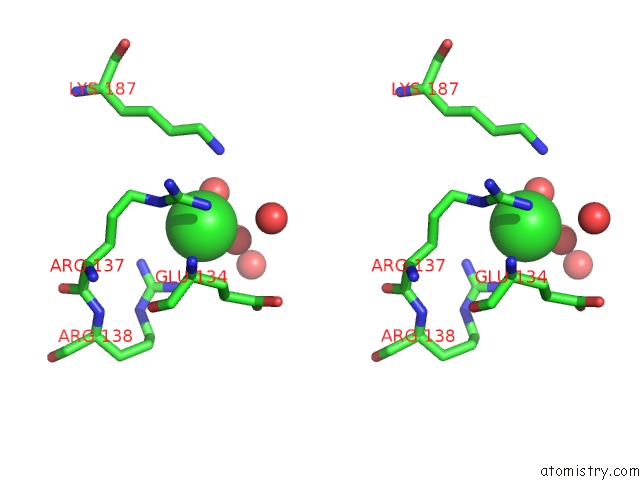
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 9 of Structure of the Plp Synthase Complex PDX1/2 (Yaad/E) From Bacillus Subtilis within 5.0Å range:
|
Chlorine binding site 10 out of 12 in 2nv2
Go back to
Chlorine binding site 10 out
of 12 in the Structure of the Plp Synthase Complex PDX1/2 (Yaad/E) From Bacillus Subtilis
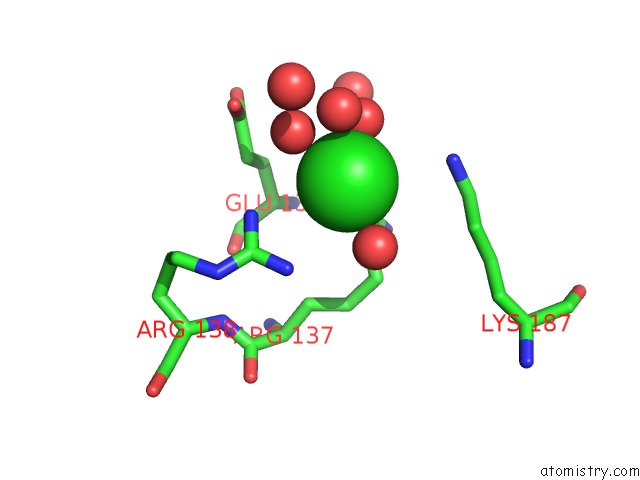
Mono view
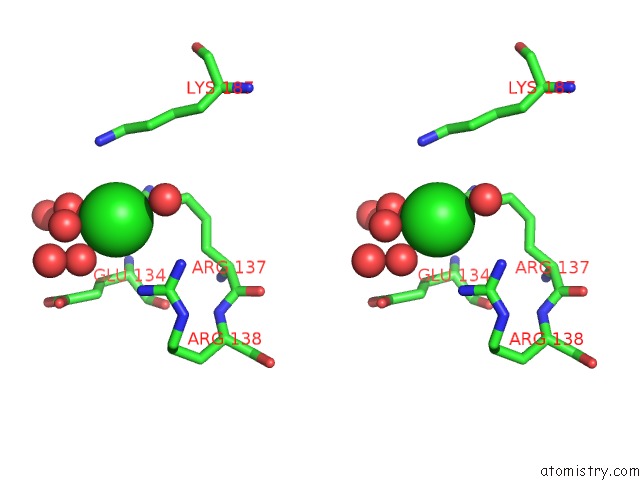
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 10 of Structure of the Plp Synthase Complex PDX1/2 (Yaad/E) From Bacillus Subtilis within 5.0Å range:
|
Reference:
M.Strohmeier,
T.Raschle,
J.Mazurkiewicz,
K.Rippe,
I.Sinning,
T.B.Fitzpatrick,
I.Tews.
Structure of A Bacterial Pyridoxal 5'-Phosphate Synthase Complex Proc.Natl.Acad.Sci.Usa V. 103 19284 2006.
ISSN: ISSN 0027-8424
PubMed: 17159152
DOI: 10.1073/PNAS.0604950103
Page generated: Thu Jul 10 23:20:39 2025
ISSN: ISSN 0027-8424
PubMed: 17159152
DOI: 10.1073/PNAS.0604950103
Last articles
Cl in 3F1VCl in 3F1S
Cl in 3F0W
Cl in 3F0A
Cl in 3F03
Cl in 3EYY
Cl in 3EZJ
Cl in 3EXN
Cl in 3EXI
Cl in 3EW5