Chlorine »
PDB 2px4-2q6r »
2q3l »
Chlorine in PDB 2q3l: Crystal Structure of An Uncharacterized Protein From DUF3478 Family with A Spoiiaa-Like Fold (SHEW_3102) From Shewanella Loihica Pv-4 at 2.25 A Resolution
Protein crystallography data
The structure of Crystal Structure of An Uncharacterized Protein From DUF3478 Family with A Spoiiaa-Like Fold (SHEW_3102) From Shewanella Loihica Pv-4 at 2.25 A Resolution, PDB code: 2q3l
was solved by
Joint Center For Structural Genomics (Jcsg),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 28.65 / 2.25 |
| Space group | C 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 40.387, 113.963, 130.692, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 18.1 / 23.9 |
Other elements in 2q3l:
The structure of Crystal Structure of An Uncharacterized Protein From DUF3478 Family with A Spoiiaa-Like Fold (SHEW_3102) From Shewanella Loihica Pv-4 at 2.25 A Resolution also contains other interesting chemical elements:
| Sodium | (Na) | 2 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of An Uncharacterized Protein From DUF3478 Family with A Spoiiaa-Like Fold (SHEW_3102) From Shewanella Loihica Pv-4 at 2.25 A Resolution
(pdb code 2q3l). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Crystal Structure of An Uncharacterized Protein From DUF3478 Family with A Spoiiaa-Like Fold (SHEW_3102) From Shewanella Loihica Pv-4 at 2.25 A Resolution, PDB code: 2q3l:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Crystal Structure of An Uncharacterized Protein From DUF3478 Family with A Spoiiaa-Like Fold (SHEW_3102) From Shewanella Loihica Pv-4 at 2.25 A Resolution, PDB code: 2q3l:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 2q3l
Go back to
Chlorine binding site 1 out
of 2 in the Crystal Structure of An Uncharacterized Protein From DUF3478 Family with A Spoiiaa-Like Fold (SHEW_3102) From Shewanella Loihica Pv-4 at 2.25 A Resolution
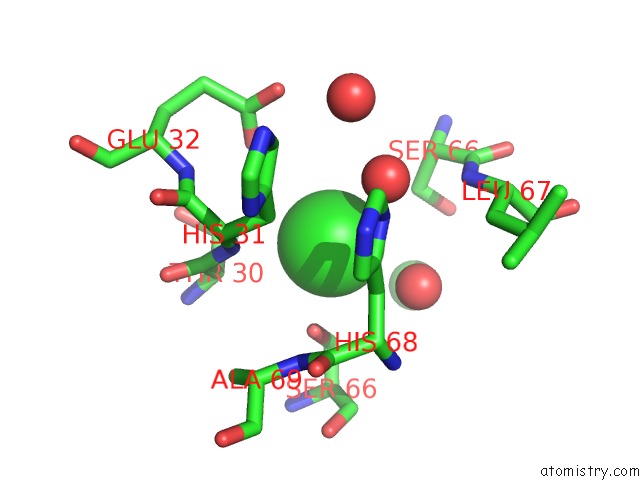
Mono view
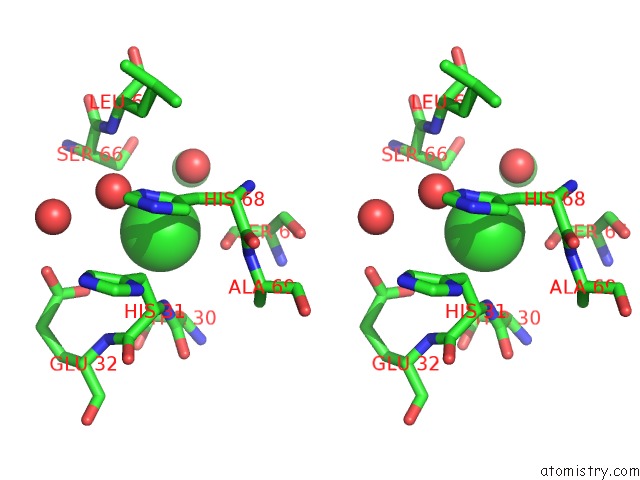
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of An Uncharacterized Protein From DUF3478 Family with A Spoiiaa-Like Fold (SHEW_3102) From Shewanella Loihica Pv-4 at 2.25 A Resolution within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 2q3l
Go back to
Chlorine binding site 2 out
of 2 in the Crystal Structure of An Uncharacterized Protein From DUF3478 Family with A Spoiiaa-Like Fold (SHEW_3102) From Shewanella Loihica Pv-4 at 2.25 A Resolution
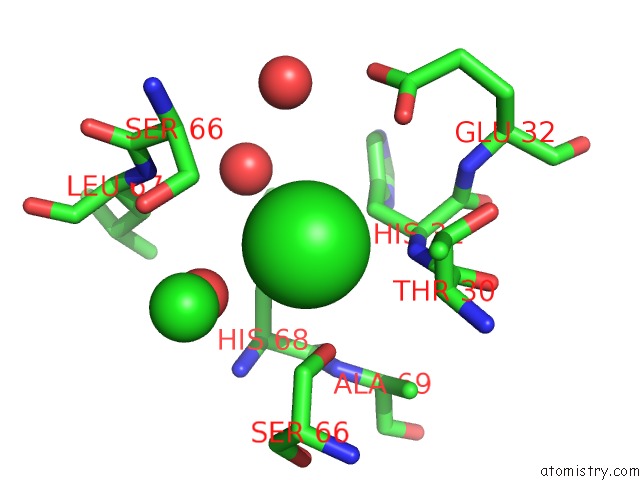
Mono view
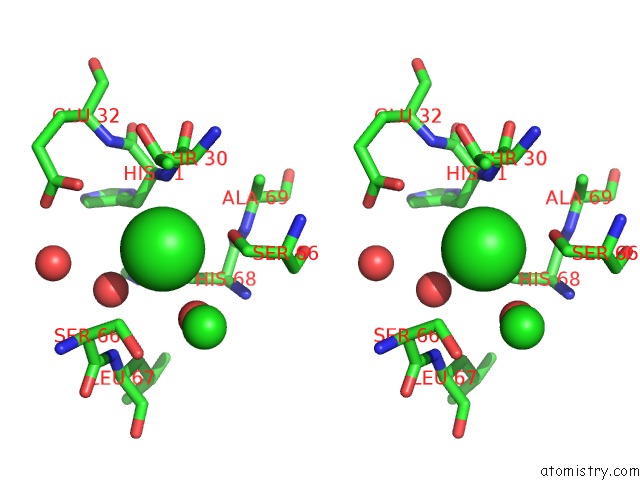
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of An Uncharacterized Protein From DUF3478 Family with A Spoiiaa-Like Fold (SHEW_3102) From Shewanella Loihica Pv-4 at 2.25 A Resolution within 5.0Å range:
|
Reference:
A.Kumar,
A.Lomize,
K.K.Jin,
D.Carlton,
M.D.Miller,
L.Jaroszewski,
P.Abdubek,
T.Astakhova,
H.L.Axelrod,
H.J.Chiu,
T.Clayton,
D.Das,
M.C.Deller,
L.Duan,
J.Feuerhelm,
J.C.Grant,
A.Grzechnik,
G.W.Han,
H.E.Klock,
M.W.Knuth,
P.Kozbial,
S.S.Krishna,
D.Marciano,
D.Mcmullan,
A.T.Morse,
E.Nigoghossian,
L.Okach,
R.Reyes,
C.L.Rife,
N.Sefcovic,
H.J.Tien,
C.B.Trame,
H.Van Den Bedem,
D.Weekes,
Q.Xu,
K.O.Hodgson,
J.Wooley,
M.A.Elsliger,
A.M.Deacon,
A.Godzik,
S.A.Lesley,
I.A.Wilson.
Open and Closed Conformations of Two Spoiiaa-Like Proteins (YP_749275.1 and YP_001095227.1) Provide Insights Into Membrane Association and Ligand Binding. Acta Crystallogr.,Sect.F V. 66 1245 2010.
ISSN: ESSN 1744-3091
PubMed: 20944218
DOI: 10.1107/S1744309109042481
Page generated: Sat Jul 20 10:25:53 2024
ISSN: ESSN 1744-3091
PubMed: 20944218
DOI: 10.1107/S1744309109042481
Last articles
Ca in 5MHCCa in 5MFL
Ca in 5MFN
Ca in 5MFH
Ca in 5MFE
Ca in 5MFG
Ca in 5MFA
Ca in 5MF4
Ca in 5MEY
Ca in 5MB1