Chlorine »
PDB 2wps-2wzc »
2ww5 »
Chlorine in PDB 2ww5: 3D-Structure of the Modular Autolysin Lytc From Streptococcus Pneumoniae at 1.6 A Resolution
Enzymatic activity of 3D-Structure of the Modular Autolysin Lytc From Streptococcus Pneumoniae at 1.6 A Resolution
All present enzymatic activity of 3D-Structure of the Modular Autolysin Lytc From Streptococcus Pneumoniae at 1.6 A Resolution:
3.2.1.17;
3.2.1.17;
Protein crystallography data
The structure of 3D-Structure of the Modular Autolysin Lytc From Streptococcus Pneumoniae at 1.6 A Resolution, PDB code: 2ww5
was solved by
I.Perez-Dorado,
R.Sanles,
J.A.Hermoso,
A.Gonzalez,
A.Garcia,
P.Garcia,
J.L.Garcia,
M.Menendez,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 72.93 / 1.61 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 59.800, 69.380, 75.870, 90.00, 106.03, 90.00 |
| R / Rfree (%) | 18.327 / 20.417 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the 3D-Structure of the Modular Autolysin Lytc From Streptococcus Pneumoniae at 1.6 A Resolution
(pdb code 2ww5). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the 3D-Structure of the Modular Autolysin Lytc From Streptococcus Pneumoniae at 1.6 A Resolution, PDB code: 2ww5:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the 3D-Structure of the Modular Autolysin Lytc From Streptococcus Pneumoniae at 1.6 A Resolution, PDB code: 2ww5:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 2ww5
Go back to
Chlorine binding site 1 out
of 2 in the 3D-Structure of the Modular Autolysin Lytc From Streptococcus Pneumoniae at 1.6 A Resolution
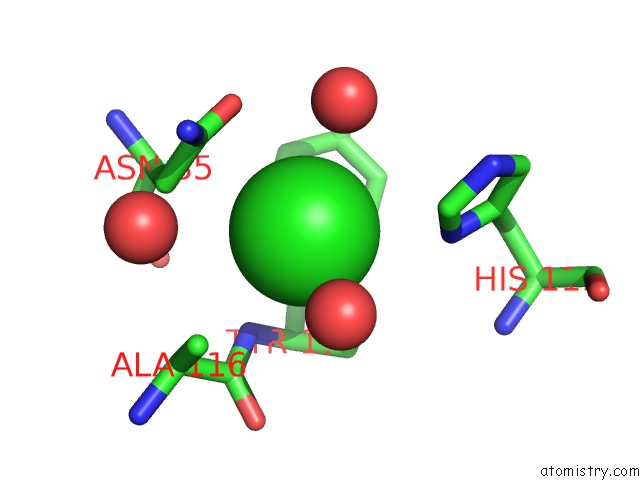
Mono view
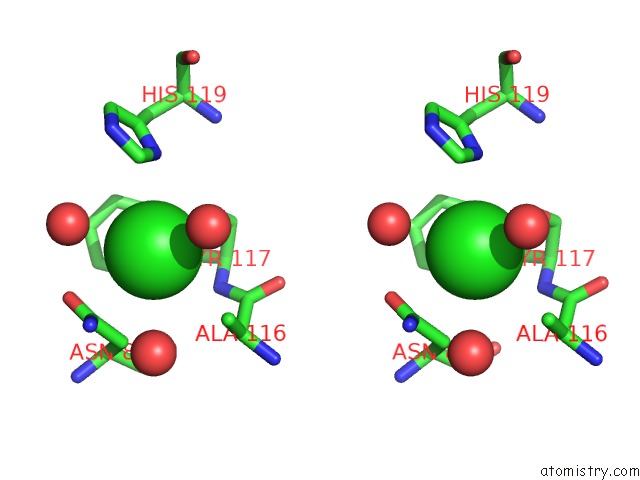
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of 3D-Structure of the Modular Autolysin Lytc From Streptococcus Pneumoniae at 1.6 A Resolution within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 2ww5
Go back to
Chlorine binding site 2 out
of 2 in the 3D-Structure of the Modular Autolysin Lytc From Streptococcus Pneumoniae at 1.6 A Resolution
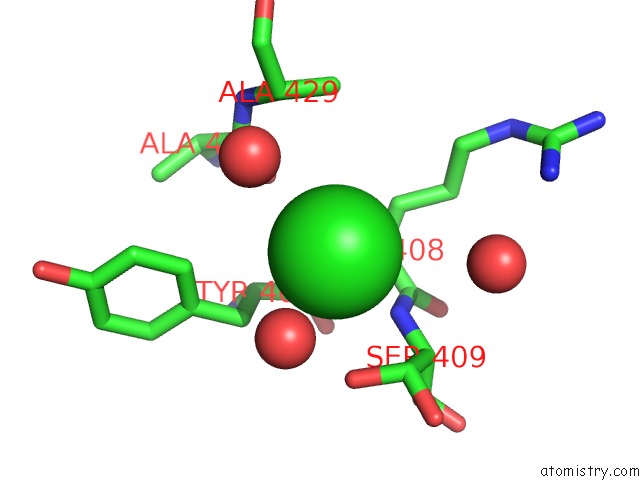
Mono view
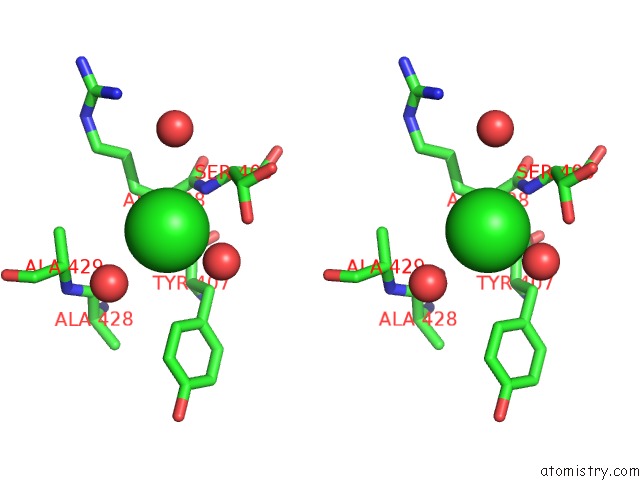
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of 3D-Structure of the Modular Autolysin Lytc From Streptococcus Pneumoniae at 1.6 A Resolution within 5.0Å range:
|
Reference:
I.Perez-Dorado,
A.Gonzalez,
M.Morales,
R.Sanles,
W.Striker,
W.Vollmer,
S.Mobashery,
J.L.Garcia,
A.Garcia,
M.Martinez-Ripoll,
P.Garcia,
J.A.Hermoso.
Insights Into Pneumococcal Fratricide From the Crystal Structures of the Modular Killing Factor Lytc. Nat.Struct.Mol.Biol. V. 17 576 2010.
ISSN: ISSN 1545-9993
PubMed: 20400948
DOI: 10.1038/NSMB.1817
Page generated: Fri Jul 11 01:42:17 2025
ISSN: ISSN 1545-9993
PubMed: 20400948
DOI: 10.1038/NSMB.1817
Last articles
Cl in 3ON7Cl in 3OOK
Cl in 3OOF
Cl in 3ONP
Cl in 3ON6
Cl in 3OMU
Cl in 3ONI
Cl in 3ON5
Cl in 3OMN
Cl in 3OMM