Chlorine »
PDB 2z0g-2zgd »
2z9x »
Chlorine in PDB 2z9x: Crystal Structure of Pyridoxamine-Pyruvate Aminotransferase Complexed with Pyridoxyl-L-Alanine
Enzymatic activity of Crystal Structure of Pyridoxamine-Pyruvate Aminotransferase Complexed with Pyridoxyl-L-Alanine
All present enzymatic activity of Crystal Structure of Pyridoxamine-Pyruvate Aminotransferase Complexed with Pyridoxyl-L-Alanine:
2.6.1.30;
2.6.1.30;
Protein crystallography data
The structure of Crystal Structure of Pyridoxamine-Pyruvate Aminotransferase Complexed with Pyridoxyl-L-Alanine, PDB code: 2z9x
was solved by
Y.Yoshikane,
N.Yokochi,
M.Yamasaki,
K.Mizutani,
K.Ohnishi,
B.Mikami,
H.Hayashi,
T.Yagi,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 14.92 / 1.94 |
| Space group | P 43 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 68.720, 68.720, 311.942, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 14.5 / 18.4 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of Pyridoxamine-Pyruvate Aminotransferase Complexed with Pyridoxyl-L-Alanine
(pdb code 2z9x). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Crystal Structure of Pyridoxamine-Pyruvate Aminotransferase Complexed with Pyridoxyl-L-Alanine, PDB code: 2z9x:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Crystal Structure of Pyridoxamine-Pyruvate Aminotransferase Complexed with Pyridoxyl-L-Alanine, PDB code: 2z9x:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 2z9x
Go back to
Chlorine binding site 1 out
of 2 in the Crystal Structure of Pyridoxamine-Pyruvate Aminotransferase Complexed with Pyridoxyl-L-Alanine
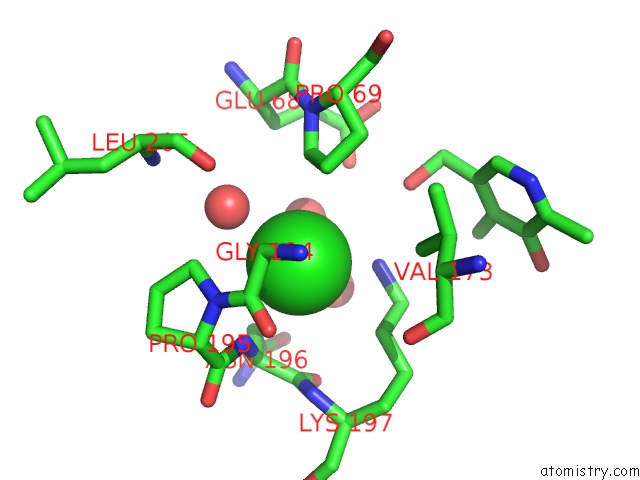
Mono view
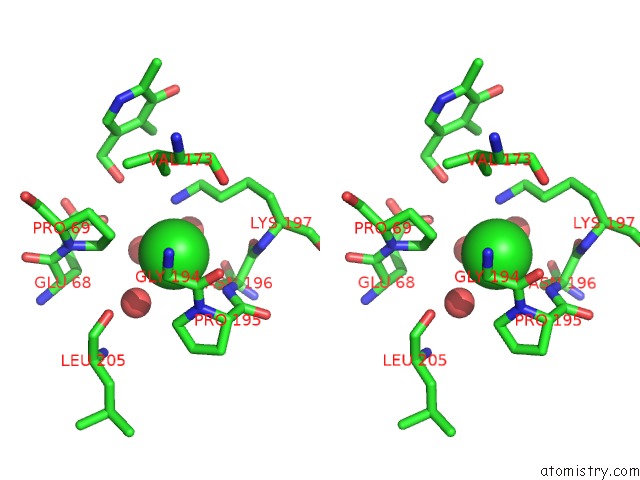
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of Pyridoxamine-Pyruvate Aminotransferase Complexed with Pyridoxyl-L-Alanine within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 2z9x
Go back to
Chlorine binding site 2 out
of 2 in the Crystal Structure of Pyridoxamine-Pyruvate Aminotransferase Complexed with Pyridoxyl-L-Alanine

Mono view
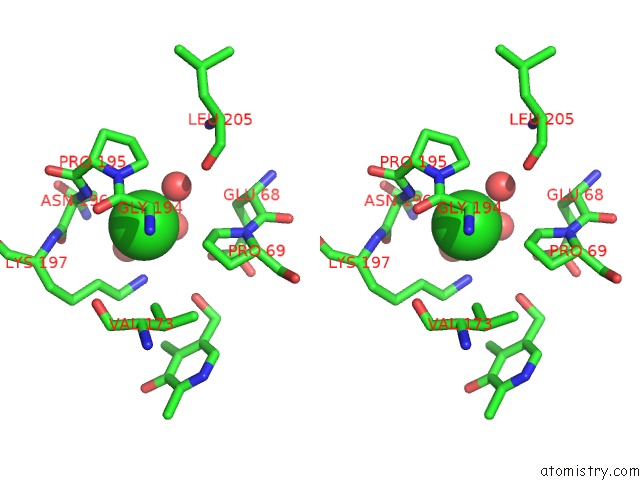
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of Pyridoxamine-Pyruvate Aminotransferase Complexed with Pyridoxyl-L-Alanine within 5.0Å range:
|
Reference:
Y.Yoshikane,
N.Yokochi,
M.Yamasaki,
K.Mizutani,
K.Ohnishi,
B.Mikami,
H.Hayashi,
T.Yagi.
Crystal Structure of Pyridoxamine-Pyruvate Aminotransferase From Mesorhizobium Loti MAFF303099 J.Biol.Chem. V. 283 1120 2008.
ISSN: ISSN 0021-9258
PubMed: 17989071
DOI: 10.1074/JBC.M708061200
Page generated: Fri Jul 11 02:52:08 2025
ISSN: ISSN 0021-9258
PubMed: 17989071
DOI: 10.1074/JBC.M708061200
Last articles
Cl in 3QDDCl in 3QCK
Cl in 3QCJ
Cl in 3QCE
Cl in 3QCF
Cl in 3QCI
Cl in 3QCH
Cl in 3QCG
Cl in 3QBF
Cl in 3QBN