Chlorine »
PDB 3ecg-3es1 »
3egk »
Chlorine in PDB 3egk: Knoble Inhibitor
Enzymatic activity of Knoble Inhibitor
All present enzymatic activity of Knoble Inhibitor:
3.4.21.5;
3.4.21.5;
Protein crystallography data
The structure of Knoble Inhibitor, PDB code: 3egk
was solved by
B.Baum,
A.Heine,
G.Klebe,
M.Muenzel,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 10.00 / 2.20 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 70.200, 71.430, 72.580, 90.00, 100.52, 90.00 |
| R / Rfree (%) | 20.5 / 31.2 |
Other elements in 3egk:
The structure of Knoble Inhibitor also contains other interesting chemical elements:
| Sodium | (Na) | 2 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Knoble Inhibitor
(pdb code 3egk). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the Knoble Inhibitor, PDB code: 3egk:
In total only one binding site of Chlorine was determined in the Knoble Inhibitor, PDB code: 3egk:
Chlorine binding site 1 out of 1 in 3egk
Go back to
Chlorine binding site 1 out
of 1 in the Knoble Inhibitor
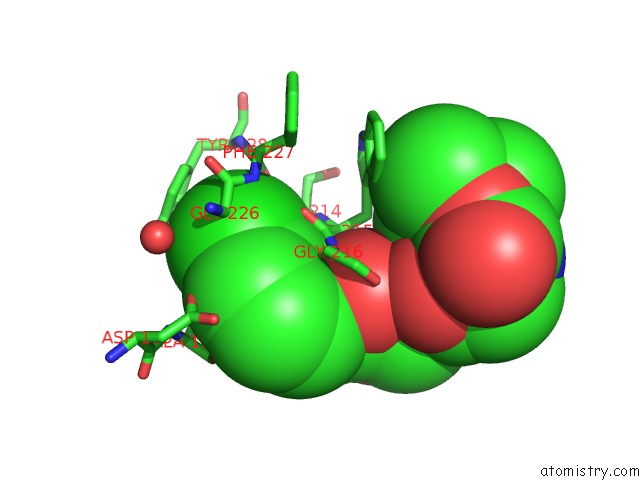
Mono view
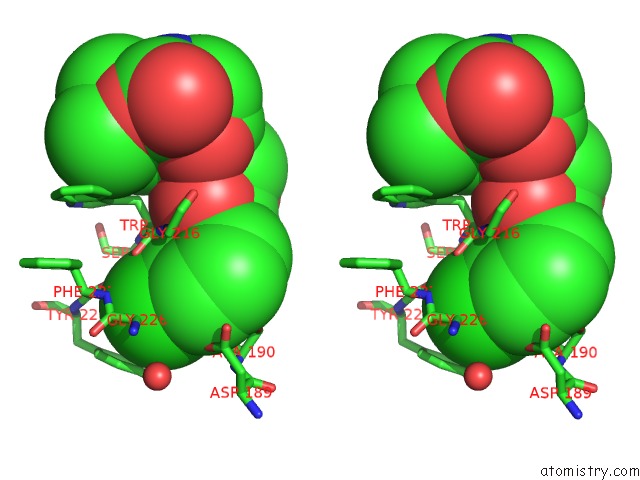
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Knoble Inhibitor within 5.0Å range:
|
Reference:
C.Gerlach,
M.Munzel,
B.Baum,
H.-D.Gerber,
T.Craan,
W.E.Diederich,
G.Klebe.
Knoble: A Knowledge-Based Approach For the Design and Synthesis of Readily Accessible Small-Molecule Chemical Probes to Test Protein Binding Angew.Chem.Int.Ed.Engl. V. 46 9105 2007.
ISSN: ISSN 1433-7851
PubMed: 17955562
DOI: 10.1002/ANIE.200703323
Page generated: Sat Jul 20 18:50:06 2024
ISSN: ISSN 1433-7851
PubMed: 17955562
DOI: 10.1002/ANIE.200703323
Last articles
Cl in 2WLKCl in 2WLH
Cl in 2WL7
Cl in 2WKY
Cl in 2WKX
Cl in 2WL5
Cl in 2WKQ
Cl in 2WKR
Cl in 2WKT
Cl in 2WKM