Chlorine »
PDB 3lz1-3m98 »
3m81 »
Chlorine in PDB 3m81: Crystal Structure of Acetyl Xylan Esterase (TM0077) From Thermotoga Maritima at 2.50 A Resolution (Native Apo Structure)
Protein crystallography data
The structure of Crystal Structure of Acetyl Xylan Esterase (TM0077) From Thermotoga Maritima at 2.50 A Resolution (Native Apo Structure), PDB code: 3m81
was solved by
Joint Center For Structural Genomics (Jcsg),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 48.80 / 2.50 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 103.460, 103.790, 221.020, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 16.7 / 21.2 |
Other elements in 3m81:
The structure of Crystal Structure of Acetyl Xylan Esterase (TM0077) From Thermotoga Maritima at 2.50 A Resolution (Native Apo Structure) also contains other interesting chemical elements:
| Calcium | (Ca) | 8 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of Acetyl Xylan Esterase (TM0077) From Thermotoga Maritima at 2.50 A Resolution (Native Apo Structure)
(pdb code 3m81). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 6 binding sites of Chlorine where determined in the Crystal Structure of Acetyl Xylan Esterase (TM0077) From Thermotoga Maritima at 2.50 A Resolution (Native Apo Structure), PDB code: 3m81:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Chlorine where determined in the Crystal Structure of Acetyl Xylan Esterase (TM0077) From Thermotoga Maritima at 2.50 A Resolution (Native Apo Structure), PDB code: 3m81:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6;
Chlorine binding site 1 out of 6 in 3m81
Go back to
Chlorine binding site 1 out
of 6 in the Crystal Structure of Acetyl Xylan Esterase (TM0077) From Thermotoga Maritima at 2.50 A Resolution (Native Apo Structure)
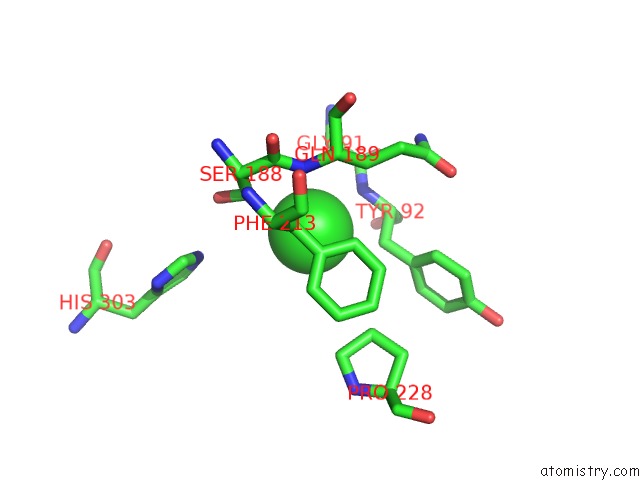
Mono view
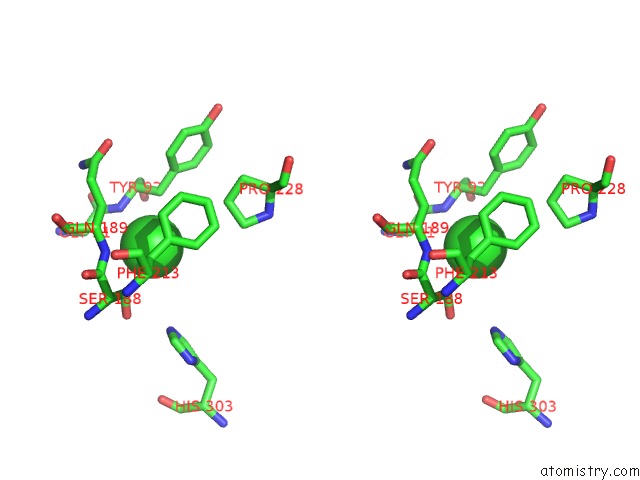
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of Acetyl Xylan Esterase (TM0077) From Thermotoga Maritima at 2.50 A Resolution (Native Apo Structure) within 5.0Å range:
|
Chlorine binding site 2 out of 6 in 3m81
Go back to
Chlorine binding site 2 out
of 6 in the Crystal Structure of Acetyl Xylan Esterase (TM0077) From Thermotoga Maritima at 2.50 A Resolution (Native Apo Structure)
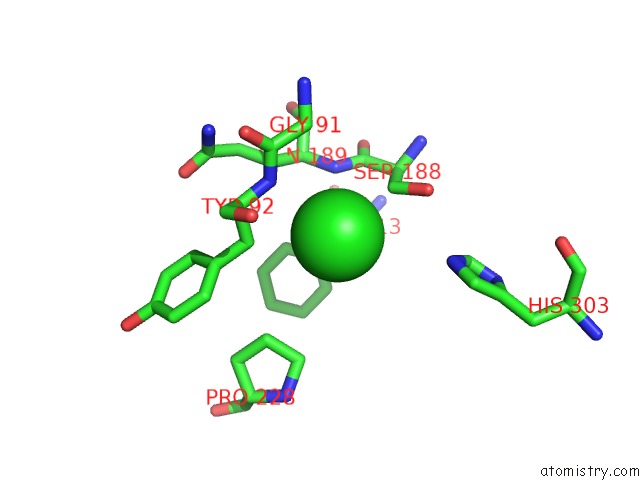
Mono view
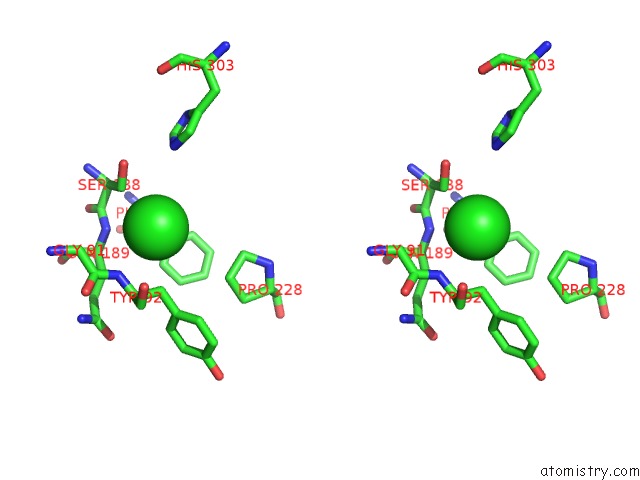
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of Acetyl Xylan Esterase (TM0077) From Thermotoga Maritima at 2.50 A Resolution (Native Apo Structure) within 5.0Å range:
|
Chlorine binding site 3 out of 6 in 3m81
Go back to
Chlorine binding site 3 out
of 6 in the Crystal Structure of Acetyl Xylan Esterase (TM0077) From Thermotoga Maritima at 2.50 A Resolution (Native Apo Structure)
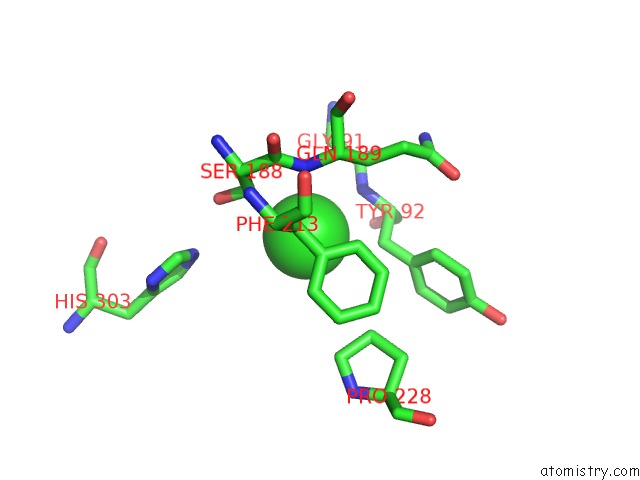
Mono view
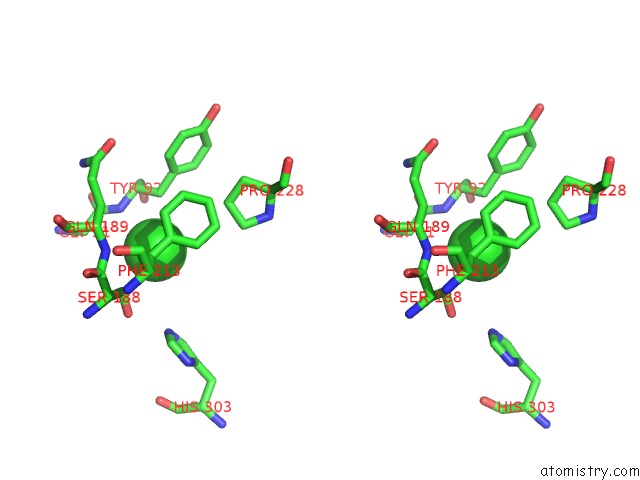
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Crystal Structure of Acetyl Xylan Esterase (TM0077) From Thermotoga Maritima at 2.50 A Resolution (Native Apo Structure) within 5.0Å range:
|
Chlorine binding site 4 out of 6 in 3m81
Go back to
Chlorine binding site 4 out
of 6 in the Crystal Structure of Acetyl Xylan Esterase (TM0077) From Thermotoga Maritima at 2.50 A Resolution (Native Apo Structure)
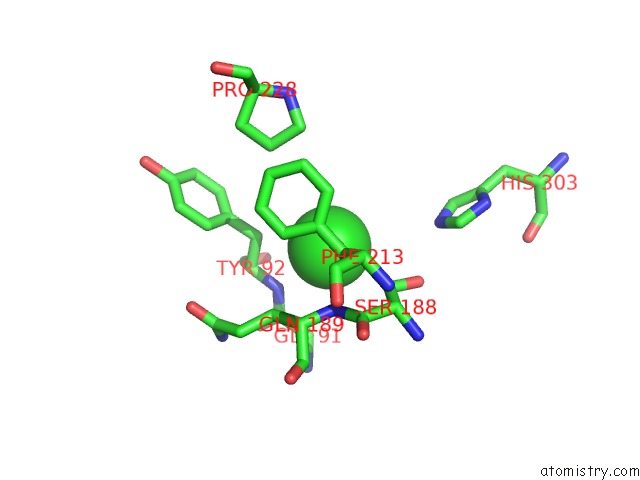
Mono view
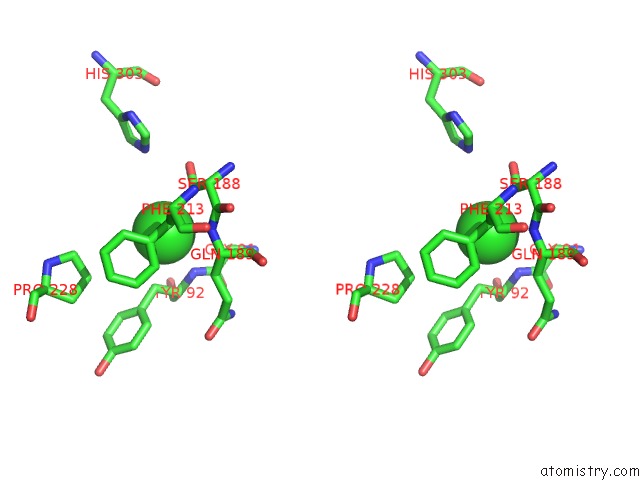
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Crystal Structure of Acetyl Xylan Esterase (TM0077) From Thermotoga Maritima at 2.50 A Resolution (Native Apo Structure) within 5.0Å range:
|
Chlorine binding site 5 out of 6 in 3m81
Go back to
Chlorine binding site 5 out
of 6 in the Crystal Structure of Acetyl Xylan Esterase (TM0077) From Thermotoga Maritima at 2.50 A Resolution (Native Apo Structure)
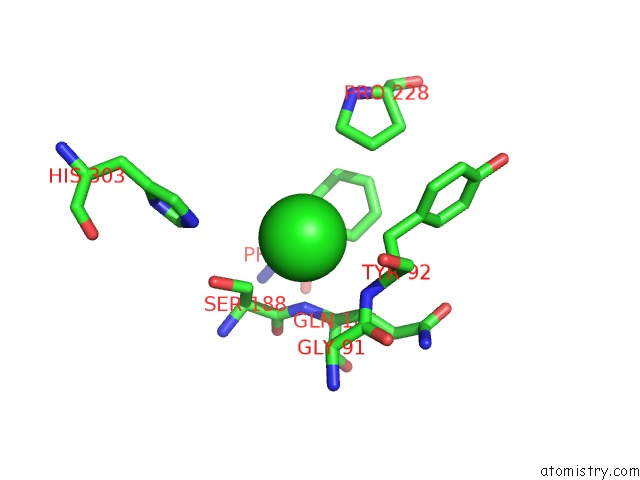
Mono view
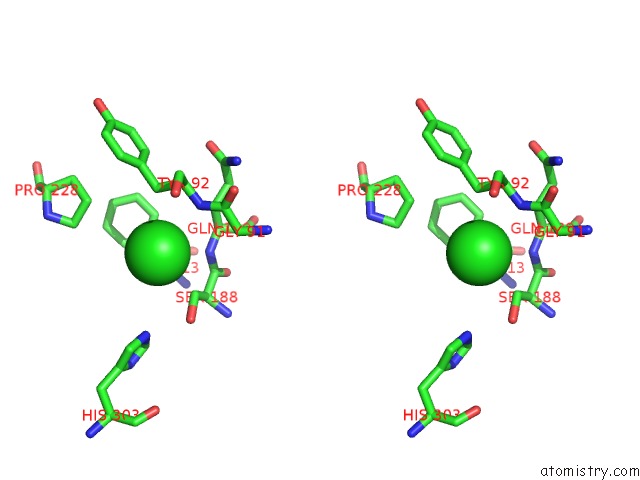
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Crystal Structure of Acetyl Xylan Esterase (TM0077) From Thermotoga Maritima at 2.50 A Resolution (Native Apo Structure) within 5.0Å range:
|
Chlorine binding site 6 out of 6 in 3m81
Go back to
Chlorine binding site 6 out
of 6 in the Crystal Structure of Acetyl Xylan Esterase (TM0077) From Thermotoga Maritima at 2.50 A Resolution (Native Apo Structure)
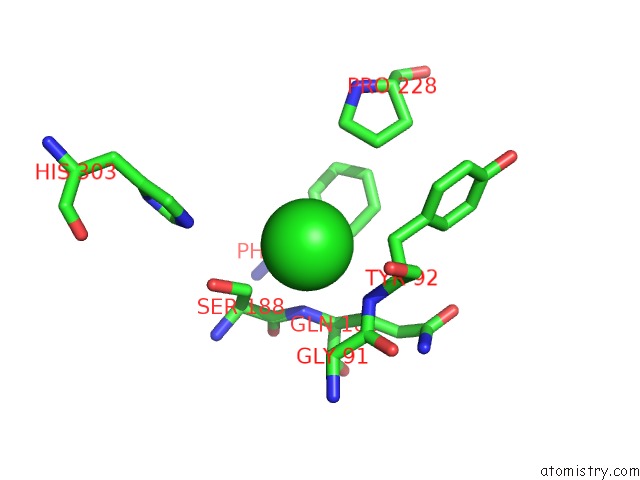
Mono view
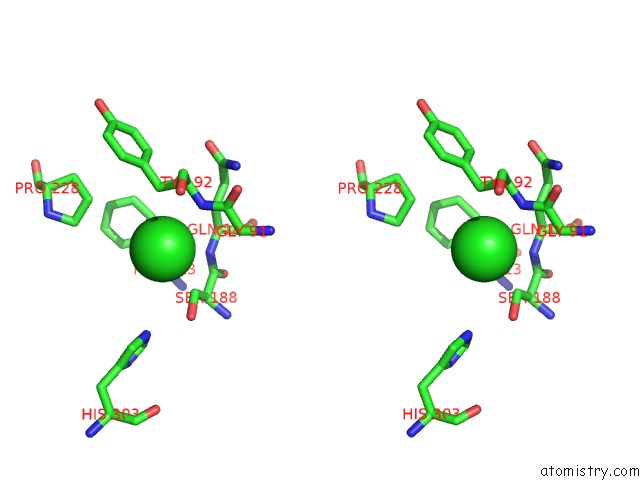
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 6 of Crystal Structure of Acetyl Xylan Esterase (TM0077) From Thermotoga Maritima at 2.50 A Resolution (Native Apo Structure) within 5.0Å range:
|
Reference:
M.Levisson,
G.W.Han,
M.C.Deller,
Q.Xu,
P.Biely,
S.Hendriks,
L.F.Ten Eyck,
C.Flensburg,
P.Roversi,
M.D.Miller,
D.Mcmullan,
F.Von Delft,
A.Kreusch,
A.M.Deacon,
J.Van Der Oost,
S.A.Lesley,
M.A.Elsliger,
S.W.Kengen,
I.A.Wilson.
Functional and Structural Characterization of A Thermostable Acetyl Esterase From Thermotoga Maritima. Proteins V. 80 1545 2012.
ISSN: ISSN 0887-3585
PubMed: 22411095
DOI: 10.1002/PROT.24041
Page generated: Fri Jul 11 07:47:05 2025
ISSN: ISSN 0887-3585
PubMed: 22411095
DOI: 10.1002/PROT.24041
Last articles
F in 4LI7F in 4LGT
F in 4LA0
F in 4LBS
F in 4LB4
F in 4LBR
F in 4L7S
F in 4LBD
F in 4LB3
F in 4LA6