Chlorine »
PDB 3qvp-3r40 »
3r0w »
Chlorine in PDB 3r0w: Crystal Structures of Multidrug-Resistant Hiv-1 Protease in Complex with Mechanism-Based Aspartyl Protease Inhibitors.
Protein crystallography data
The structure of Crystal Structures of Multidrug-Resistant Hiv-1 Protease in Complex with Mechanism-Based Aspartyl Protease Inhibitors., PDB code: 3r0w
was solved by
R.S.Yedidi,
D.Gupta,
Z.Liu,
J.Brunzelle,
I.A.Kovari,
P.M.Woster,
L.C.Kovari,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 18.98 / 1.70 |
| Space group | P 41 |
| Cell size a, b, c (Å), α, β, γ (°) | 45.727, 45.727, 102.149, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 19.8 / 24.5 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structures of Multidrug-Resistant Hiv-1 Protease in Complex with Mechanism-Based Aspartyl Protease Inhibitors.
(pdb code 3r0w). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the Crystal Structures of Multidrug-Resistant Hiv-1 Protease in Complex with Mechanism-Based Aspartyl Protease Inhibitors., PDB code: 3r0w:
In total only one binding site of Chlorine was determined in the Crystal Structures of Multidrug-Resistant Hiv-1 Protease in Complex with Mechanism-Based Aspartyl Protease Inhibitors., PDB code: 3r0w:
Chlorine binding site 1 out of 1 in 3r0w
Go back to
Chlorine binding site 1 out
of 1 in the Crystal Structures of Multidrug-Resistant Hiv-1 Protease in Complex with Mechanism-Based Aspartyl Protease Inhibitors.
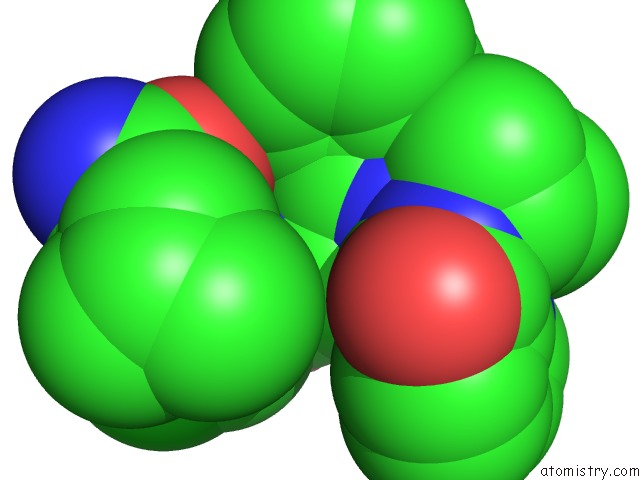
Mono view
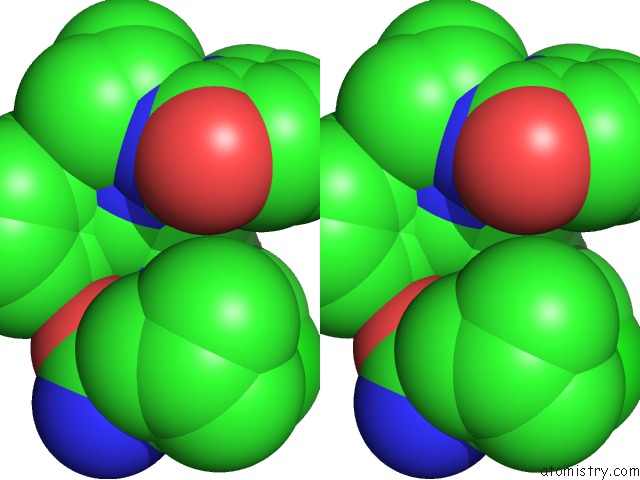
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structures of Multidrug-Resistant Hiv-1 Protease in Complex with Mechanism-Based Aspartyl Protease Inhibitors. within 5.0Å range:
|
Reference:
R.S.Yedidi,
Z.Liu,
Y.Wang,
J.S.Brunzelle,
I.A.Kovari,
P.M.Woster,
L.C.Kovari,
D.Gupta.
Crystal Structures of Multidrug-Resistant Hiv-1 Protease in Complex with Two Potent Anti-Malarial Compounds. Biochem.Biophys.Res.Commun. V. 421 413 2012.
ISSN: ISSN 0006-291X
PubMed: 22469467
DOI: 10.1016/J.BBRC.2012.03.096
Page generated: Sun Jul 21 03:20:35 2024
ISSN: ISSN 0006-291X
PubMed: 22469467
DOI: 10.1016/J.BBRC.2012.03.096
Last articles
Ca in 5TGQCa in 5TGF
Ca in 5TFM
Ca in 5TEA
Ca in 5TFL
Ca in 5TD4
Ca in 5TF9
Ca in 5TAK
Ca in 5TAE
Ca in 5TAJ