Chlorine »
PDB 3r41-3rcg »
3rbu »
Chlorine in PDB 3rbu: N-Terminally Avitev-Tagged Human Glutamate Carboxypeptidase II in Complex with 2-Pmpa
Enzymatic activity of N-Terminally Avitev-Tagged Human Glutamate Carboxypeptidase II in Complex with 2-Pmpa
All present enzymatic activity of N-Terminally Avitev-Tagged Human Glutamate Carboxypeptidase II in Complex with 2-Pmpa:
3.4.17.21;
3.4.17.21;
Protein crystallography data
The structure of N-Terminally Avitev-Tagged Human Glutamate Carboxypeptidase II in Complex with 2-Pmpa, PDB code: 3rbu
was solved by
J.Tykvart,
P.Sacha,
C.Barinka,
J.Starkova,
T.Knedlik,
J.Lubkowski,
J.Konvalinka,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 28.46 / 1.60 |
| Space group | I 2 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 101.247, 130.559, 158.897, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 15.7 / 18.2 |
Other elements in 3rbu:
The structure of N-Terminally Avitev-Tagged Human Glutamate Carboxypeptidase II in Complex with 2-Pmpa also contains other interesting chemical elements:
| Zinc | (Zn) | 2 atoms |
| Calcium | (Ca) | 1 atom |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the N-Terminally Avitev-Tagged Human Glutamate Carboxypeptidase II in Complex with 2-Pmpa
(pdb code 3rbu). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the N-Terminally Avitev-Tagged Human Glutamate Carboxypeptidase II in Complex with 2-Pmpa, PDB code: 3rbu:
In total only one binding site of Chlorine was determined in the N-Terminally Avitev-Tagged Human Glutamate Carboxypeptidase II in Complex with 2-Pmpa, PDB code: 3rbu:
Chlorine binding site 1 out of 1 in 3rbu
Go back to
Chlorine binding site 1 out
of 1 in the N-Terminally Avitev-Tagged Human Glutamate Carboxypeptidase II in Complex with 2-Pmpa
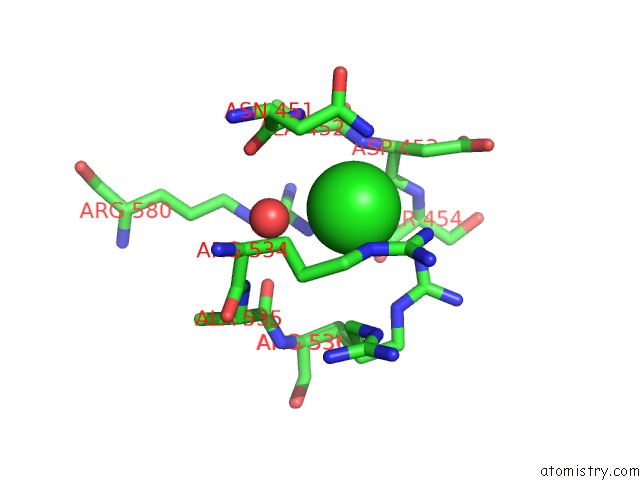
Mono view
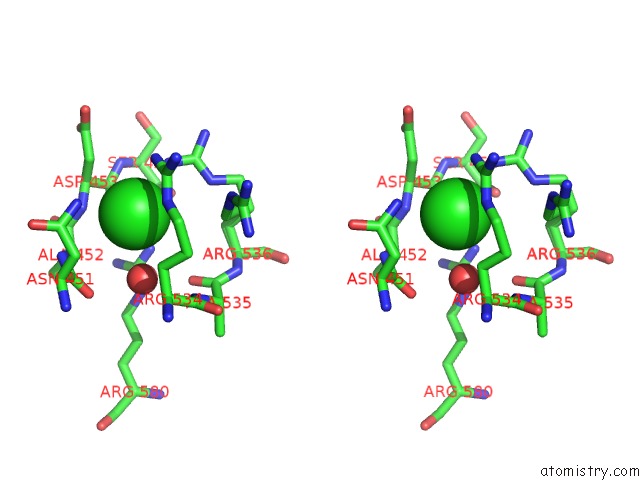
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of N-Terminally Avitev-Tagged Human Glutamate Carboxypeptidase II in Complex with 2-Pmpa within 5.0Å range:
|
Reference:
J.Tykvart,
P.Sacha,
C.Barinka,
T.Knedlik,
J.Starkova,
J.Lubkowski,
J.Konvalinka.
Efficient and Versatile One-Step Affinity Purification of in Vivo Biotinylated Proteins: Expression, Characterization and Structure Analysis of Recombinant Human Glutamate Carboxypeptidase II. Protein Expr.Purif. V. 82 106 2012.
ISSN: ISSN 1046-5928
PubMed: 22178733
DOI: 10.1016/J.PEP.2011.11.016
Page generated: Fri Jul 11 09:47:19 2025
ISSN: ISSN 1046-5928
PubMed: 22178733
DOI: 10.1016/J.PEP.2011.11.016
Last articles
F in 4MDDF in 4MDB
F in 4MC9
F in 4MDA
F in 4MC6
F in 4MC2
F in 4MC1
F in 4M8N
F in 4MBS
F in 4MBJ