Chlorine »
PDB 3s8o-3seo »
3sds »
Chlorine in PDB 3sds: Crystal Structure of A Mitochondrial Ornithine Carbamoyltransferase From Coccidioides Immitis
Enzymatic activity of Crystal Structure of A Mitochondrial Ornithine Carbamoyltransferase From Coccidioides Immitis
All present enzymatic activity of Crystal Structure of A Mitochondrial Ornithine Carbamoyltransferase From Coccidioides Immitis:
2.1.3.3;
2.1.3.3;
Protein crystallography data
The structure of Crystal Structure of A Mitochondrial Ornithine Carbamoyltransferase From Coccidioides Immitis, PDB code: 3sds
was solved by
T.E.Edwards,
J.Abendroth,
Seattle Structural Genomics Center Forinfectious Disease (Ssgcid),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 50.00 / 2.80 |
| Space group | P 3 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 150.360, 150.360, 92.110, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 19.3 / 23.9 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of A Mitochondrial Ornithine Carbamoyltransferase From Coccidioides Immitis
(pdb code 3sds). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 6 binding sites of Chlorine where determined in the Crystal Structure of A Mitochondrial Ornithine Carbamoyltransferase From Coccidioides Immitis, PDB code: 3sds:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Chlorine where determined in the Crystal Structure of A Mitochondrial Ornithine Carbamoyltransferase From Coccidioides Immitis, PDB code: 3sds:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6;
Chlorine binding site 1 out of 6 in 3sds
Go back to
Chlorine binding site 1 out
of 6 in the Crystal Structure of A Mitochondrial Ornithine Carbamoyltransferase From Coccidioides Immitis
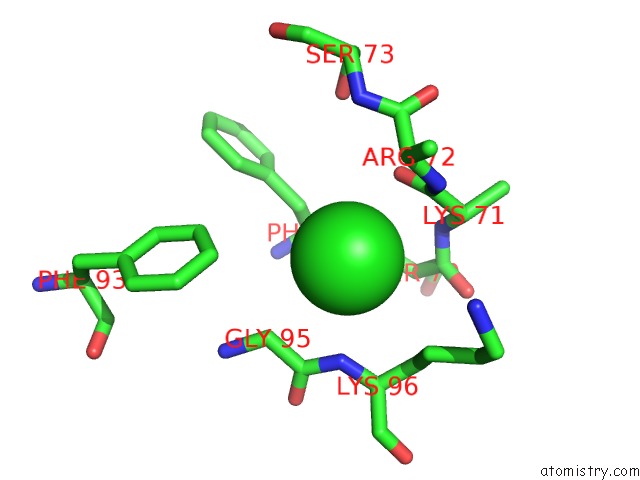
Mono view
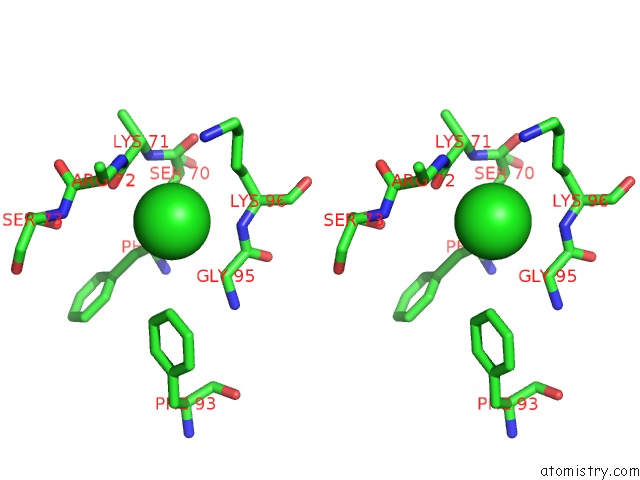
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of A Mitochondrial Ornithine Carbamoyltransferase From Coccidioides Immitis within 5.0Å range:
|
Chlorine binding site 2 out of 6 in 3sds
Go back to
Chlorine binding site 2 out
of 6 in the Crystal Structure of A Mitochondrial Ornithine Carbamoyltransferase From Coccidioides Immitis
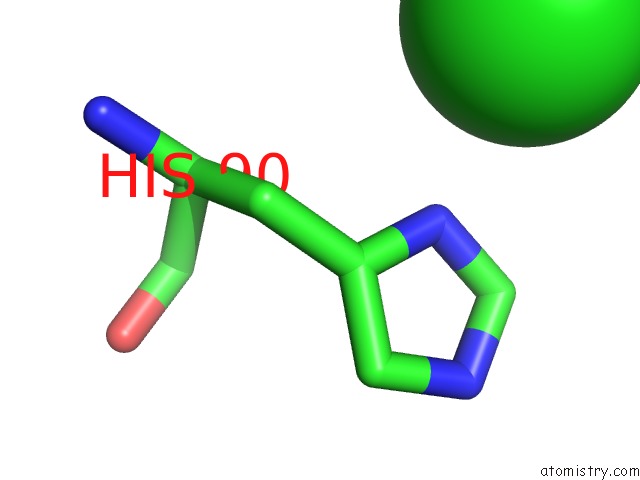
Mono view
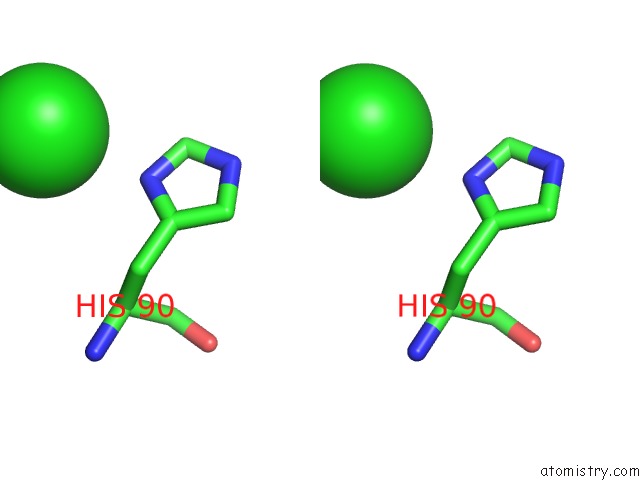
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of A Mitochondrial Ornithine Carbamoyltransferase From Coccidioides Immitis within 5.0Å range:
|
Chlorine binding site 3 out of 6 in 3sds
Go back to
Chlorine binding site 3 out
of 6 in the Crystal Structure of A Mitochondrial Ornithine Carbamoyltransferase From Coccidioides Immitis
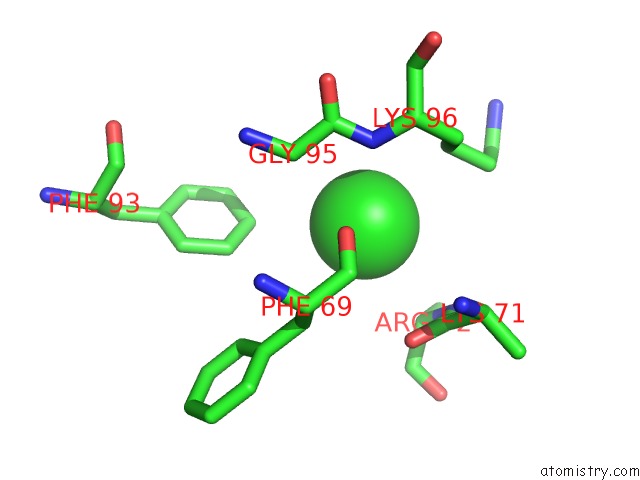
Mono view
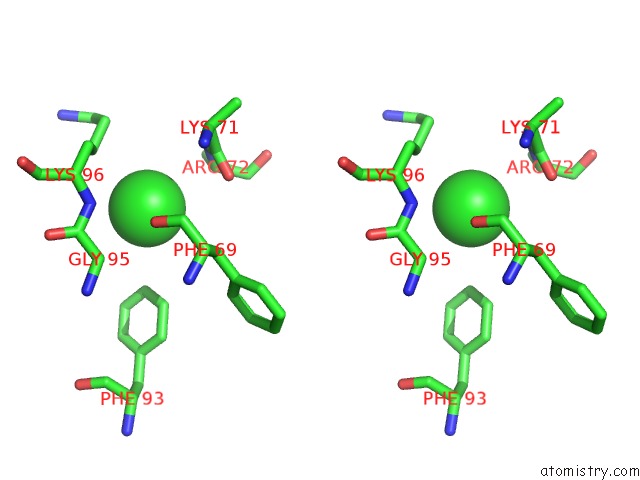
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Crystal Structure of A Mitochondrial Ornithine Carbamoyltransferase From Coccidioides Immitis within 5.0Å range:
|
Chlorine binding site 4 out of 6 in 3sds
Go back to
Chlorine binding site 4 out
of 6 in the Crystal Structure of A Mitochondrial Ornithine Carbamoyltransferase From Coccidioides Immitis
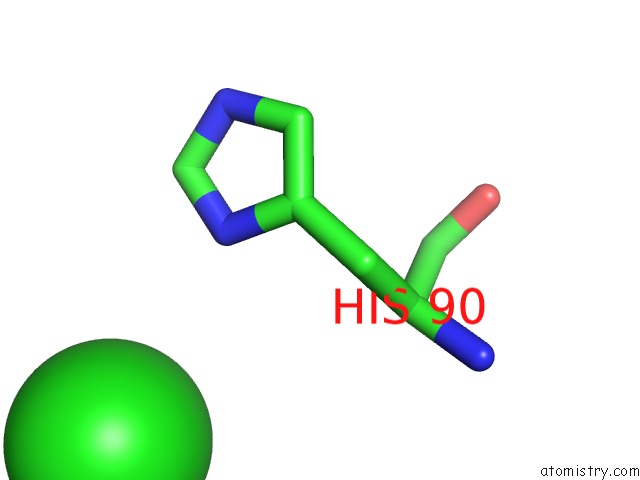
Mono view
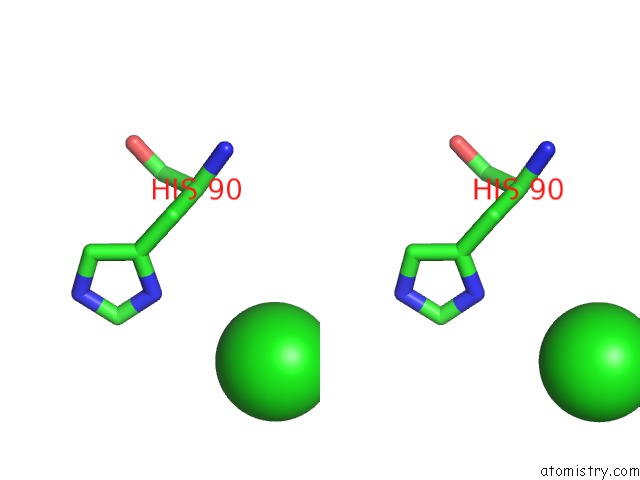
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Crystal Structure of A Mitochondrial Ornithine Carbamoyltransferase From Coccidioides Immitis within 5.0Å range:
|
Chlorine binding site 5 out of 6 in 3sds
Go back to
Chlorine binding site 5 out
of 6 in the Crystal Structure of A Mitochondrial Ornithine Carbamoyltransferase From Coccidioides Immitis
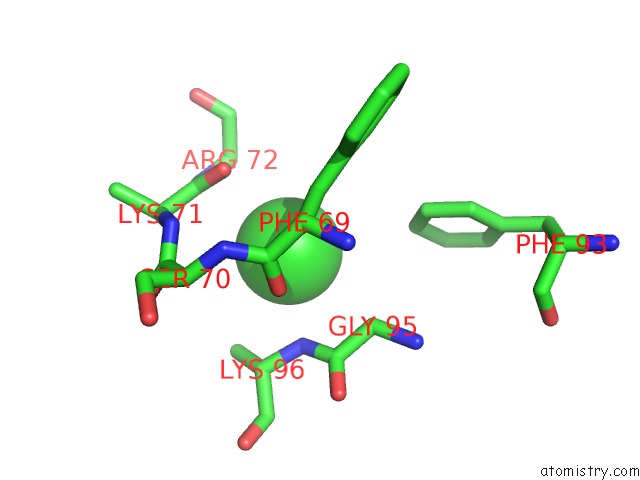
Mono view
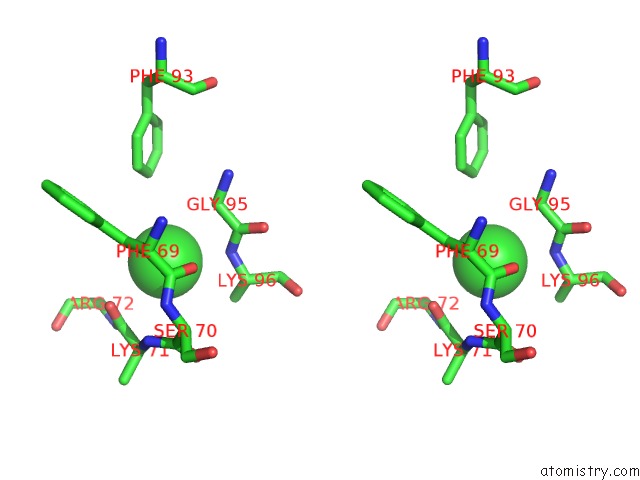
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Crystal Structure of A Mitochondrial Ornithine Carbamoyltransferase From Coccidioides Immitis within 5.0Å range:
|
Chlorine binding site 6 out of 6 in 3sds
Go back to
Chlorine binding site 6 out
of 6 in the Crystal Structure of A Mitochondrial Ornithine Carbamoyltransferase From Coccidioides Immitis
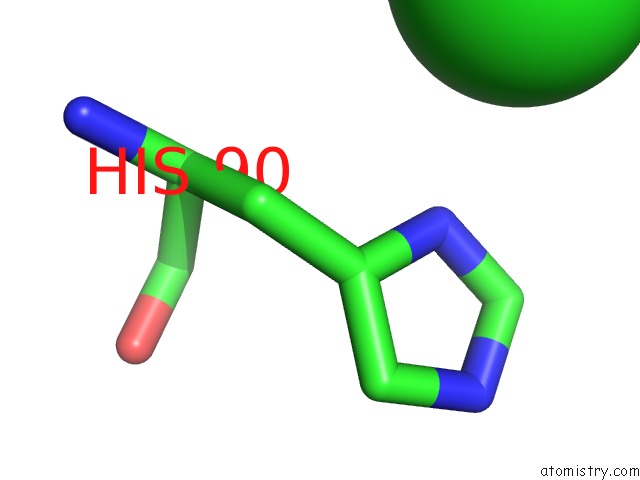
Mono view
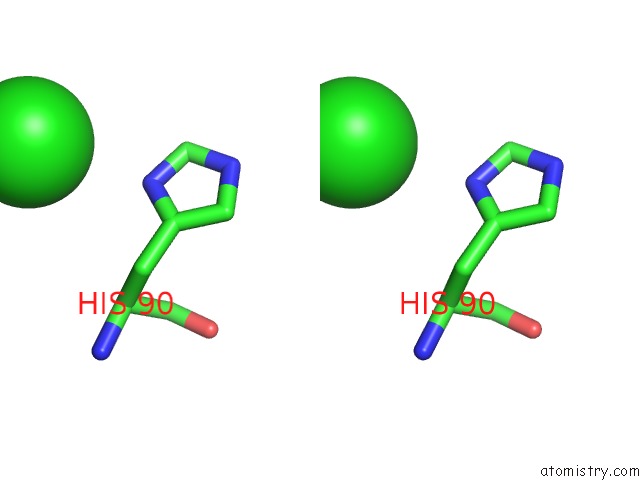
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 6 of Crystal Structure of A Mitochondrial Ornithine Carbamoyltransferase From Coccidioides Immitis within 5.0Å range:
|
Reference:
S.N.Hewitt,
R.Choi,
A.Kelley,
G.J.Crowther,
A.J.Napuli,
W.C.Van Voorhis.
Expression of Proteins in Escherichia Coli As Fusions with Maltose-Binding Protein to Rescue Non-Expressed Targets in A High-Throughput Protein-Expression and Purification Pipeline. Acta Crystallogr.,Sect.F V. 67 1006 2011.
ISSN: ESSN 1744-3091
PubMed: 21904041
DOI: 10.1107/S1744309111022159
Page generated: Sun Jul 21 04:22:32 2024
ISSN: ESSN 1744-3091
PubMed: 21904041
DOI: 10.1107/S1744309111022159
Last articles
Ca in 2RLDCa in 2RNT
Ca in 2RJQ
Ca in 2RIE
Ca in 2RID
Ca in 2RJI
Ca in 2RIC
Ca in 2RIB
Ca in 2RIA
Ca in 2RF7