Chlorine »
PDB 3txg-3u69 »
3txh »
Chlorine in PDB 3txh: Hewl Co-Crystallization with Carboplatin in Dmso Media with Glycerol As the Cryoprotectant
Enzymatic activity of Hewl Co-Crystallization with Carboplatin in Dmso Media with Glycerol As the Cryoprotectant
All present enzymatic activity of Hewl Co-Crystallization with Carboplatin in Dmso Media with Glycerol As the Cryoprotectant:
3.2.1.17;
3.2.1.17;
Protein crystallography data
The structure of Hewl Co-Crystallization with Carboplatin in Dmso Media with Glycerol As the Cryoprotectant, PDB code: 3txh
was solved by
S.W.M.Tanley,
A.M.M.Schreurs,
J.R.Helliwell,
L.M.J.Kroon-Batenburg,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 55.75 / 1.69 |
| Space group | P 43 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 78.847, 78.847, 37.036, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 16.7 / 23.3 |
Other elements in 3txh:
The structure of Hewl Co-Crystallization with Carboplatin in Dmso Media with Glycerol As the Cryoprotectant also contains other interesting chemical elements:
| Platinum | (Pt) | 2 atoms |
| Sodium | (Na) | 1 atom |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Hewl Co-Crystallization with Carboplatin in Dmso Media with Glycerol As the Cryoprotectant
(pdb code 3txh). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 7 binding sites of Chlorine where determined in the Hewl Co-Crystallization with Carboplatin in Dmso Media with Glycerol As the Cryoprotectant, PDB code: 3txh:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7;
In total 7 binding sites of Chlorine where determined in the Hewl Co-Crystallization with Carboplatin in Dmso Media with Glycerol As the Cryoprotectant, PDB code: 3txh:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7;
Chlorine binding site 1 out of 7 in 3txh
Go back to
Chlorine binding site 1 out
of 7 in the Hewl Co-Crystallization with Carboplatin in Dmso Media with Glycerol As the Cryoprotectant
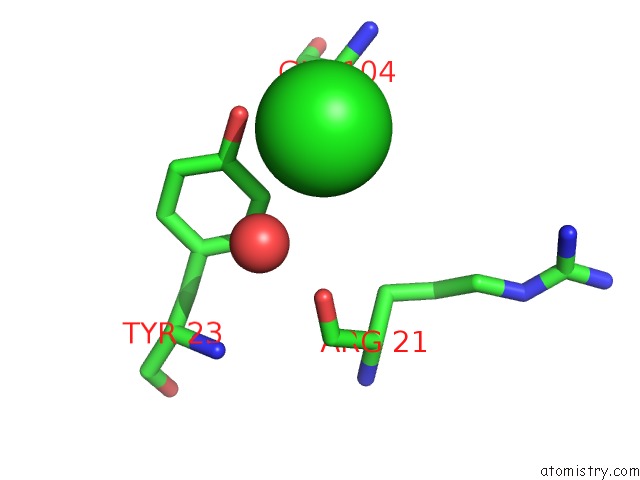
Mono view
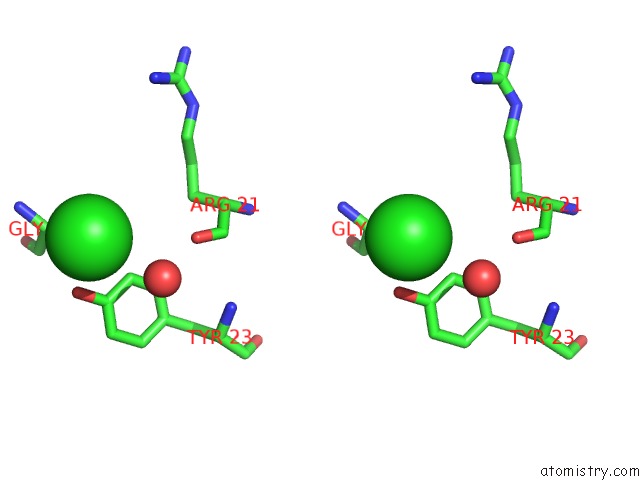
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Hewl Co-Crystallization with Carboplatin in Dmso Media with Glycerol As the Cryoprotectant within 5.0Å range:
|
Chlorine binding site 2 out of 7 in 3txh
Go back to
Chlorine binding site 2 out
of 7 in the Hewl Co-Crystallization with Carboplatin in Dmso Media with Glycerol As the Cryoprotectant
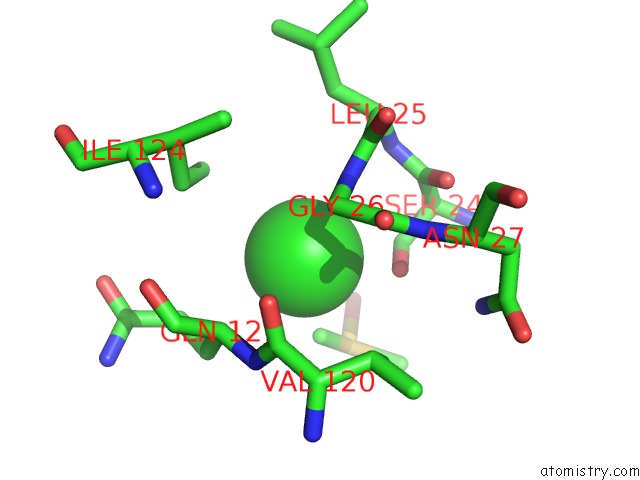
Mono view
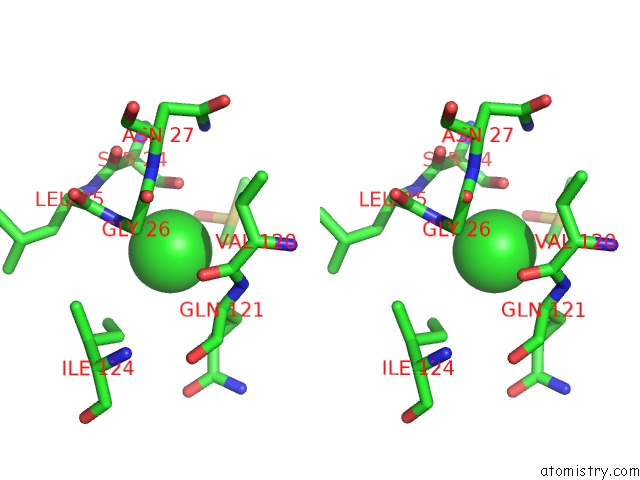
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Hewl Co-Crystallization with Carboplatin in Dmso Media with Glycerol As the Cryoprotectant within 5.0Å range:
|
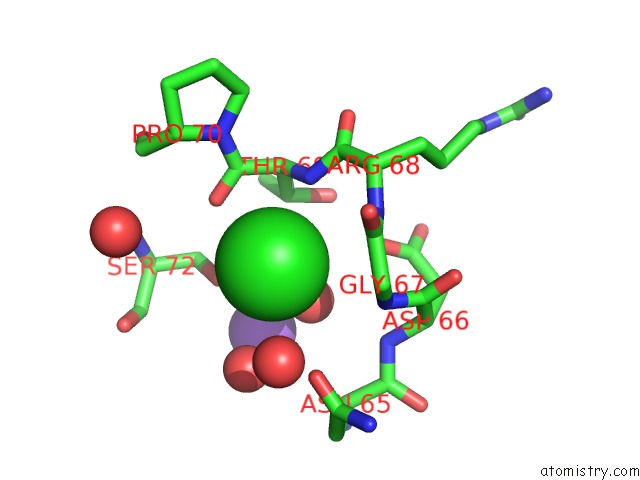
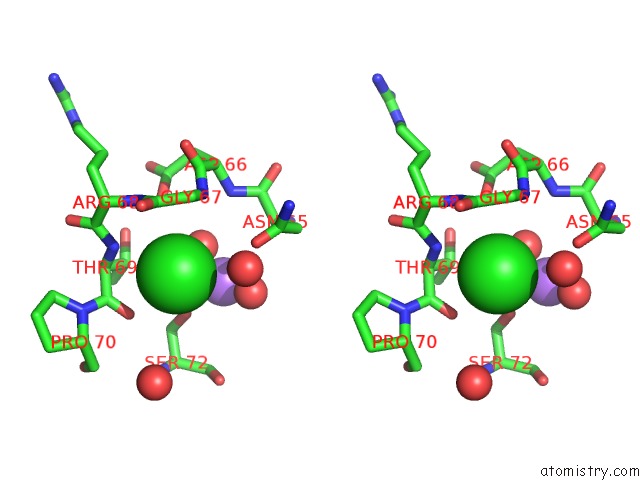
Chlorine binding site 3 out of 7 in 3txh
Go back to
Chlorine binding site 3 out
of 7 in the Hewl Co-Crystallization with Carboplatin in Dmso Media with Glycerol As the Cryoprotectant
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Hewl Co-Crystallization with Carboplatin in Dmso Media with Glycerol As the Cryoprotectant within 5.0Å range:
|
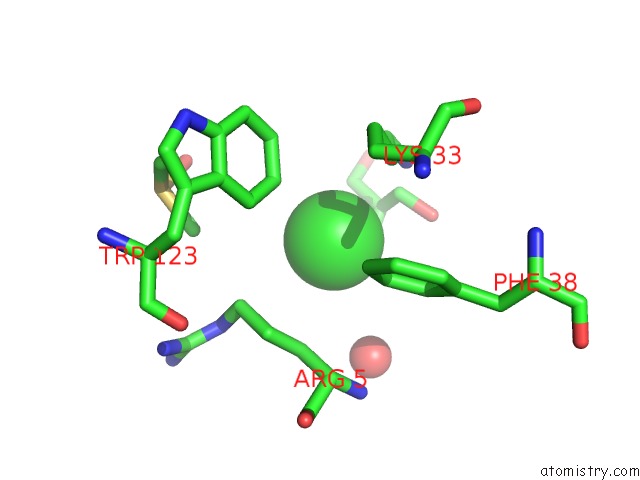
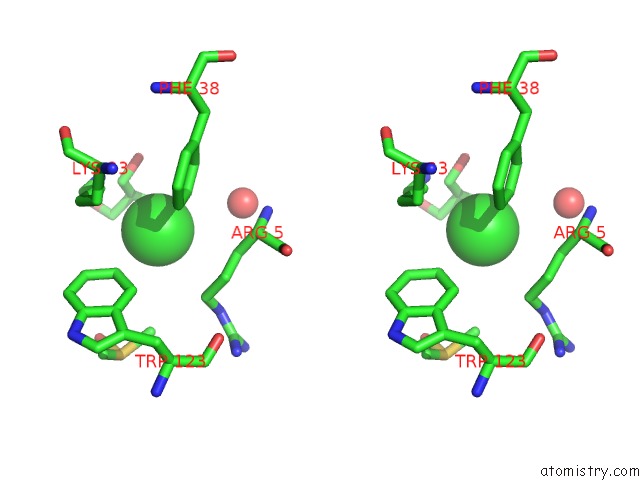
Chlorine binding site 4 out of 7 in 3txh
Go back to
Chlorine binding site 4 out
of 7 in the Hewl Co-Crystallization with Carboplatin in Dmso Media with Glycerol As the Cryoprotectant
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Hewl Co-Crystallization with Carboplatin in Dmso Media with Glycerol As the Cryoprotectant within 5.0Å range:
|
Chlorine binding site 5 out of 7 in 3txh
Go back to
Chlorine binding site 5 out
of 7 in the Hewl Co-Crystallization with Carboplatin in Dmso Media with Glycerol As the Cryoprotectant
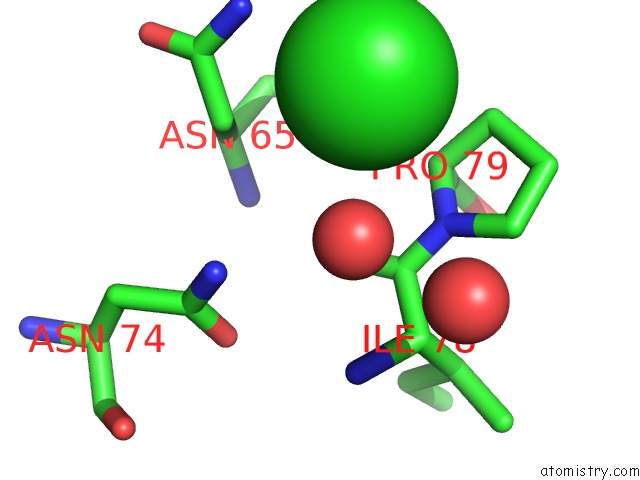
Mono view
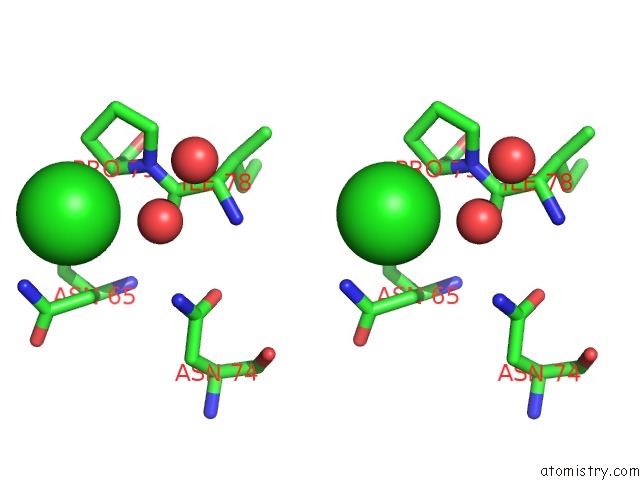
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Hewl Co-Crystallization with Carboplatin in Dmso Media with Glycerol As the Cryoprotectant within 5.0Å range:
|
Chlorine binding site 6 out of 7 in 3txh
Go back to
Chlorine binding site 6 out
of 7 in the Hewl Co-Crystallization with Carboplatin in Dmso Media with Glycerol As the Cryoprotectant
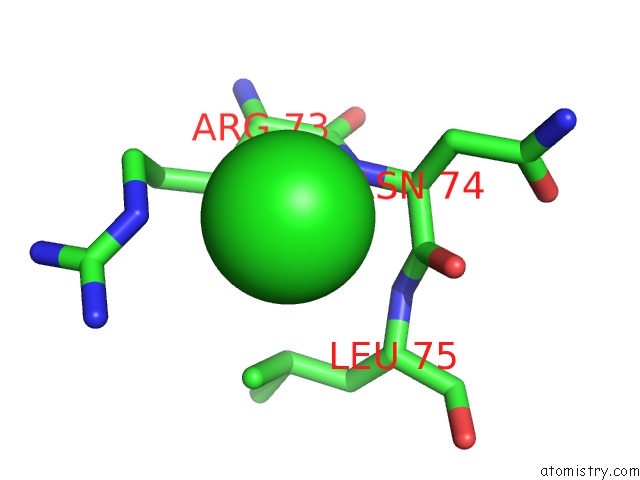
Mono view
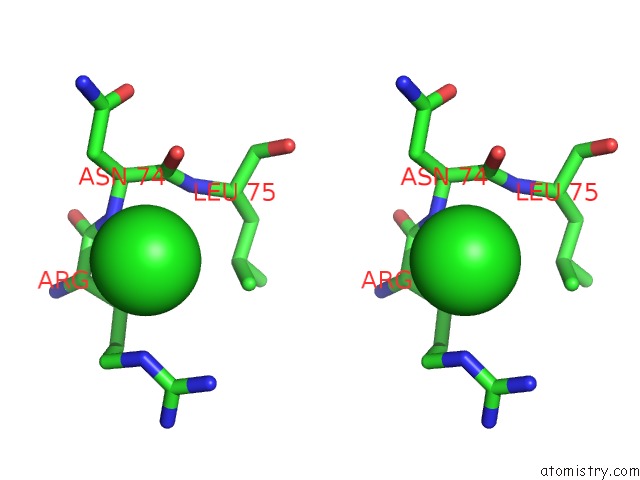
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 6 of Hewl Co-Crystallization with Carboplatin in Dmso Media with Glycerol As the Cryoprotectant within 5.0Å range:
|
Chlorine binding site 7 out of 7 in 3txh
Go back to
Chlorine binding site 7 out
of 7 in the Hewl Co-Crystallization with Carboplatin in Dmso Media with Glycerol As the Cryoprotectant
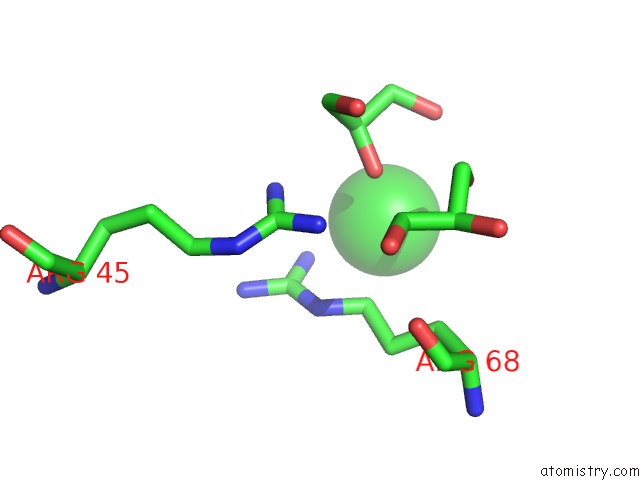
Mono view
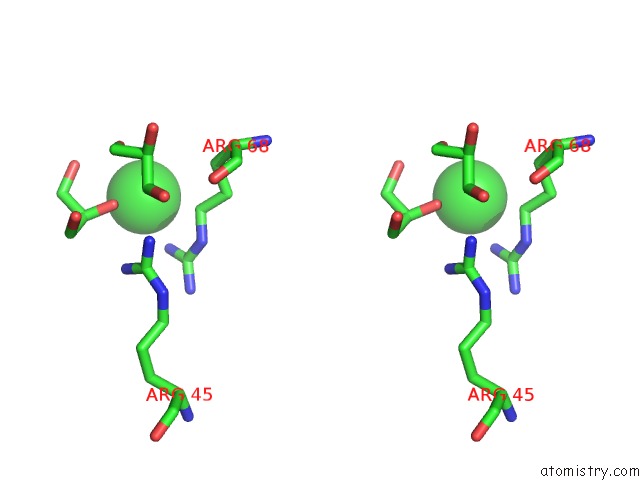
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 7 of Hewl Co-Crystallization with Carboplatin in Dmso Media with Glycerol As the Cryoprotectant within 5.0Å range:
|
Reference:
S.W.Tanley,
A.M.Schreurs,
J.R.Helliwell,
L.M.Kroon-Batenburg.
Experience with Exchange and Archiving of Raw Data: Comparison of Data From Two Diffractometers and Four Software Packages on A Series of Lysozyme Crystals. J.Appl.Crystallogr. V. 46 108 2013.
ISSN: ISSN 0021-8898
PubMed: 23396873
DOI: 10.1107/S0021889812044172
Page generated: Fri Jul 11 11:00:06 2025
ISSN: ISSN 0021-8898
PubMed: 23396873
DOI: 10.1107/S0021889812044172
Last articles
F in 4IQVF in 4IQW
F in 4IQT
F in 4IQU
F in 4INB
F in 4IKT
F in 4IJU
F in 4IKL
F in 4IJV
F in 4IKK