Chlorine »
PDB 3v6e-3vfb »
3vbg »
Chlorine in PDB 3vbg: Structure of HDM2 with Dimer Inducing Indolyl Hydantoin Ro-2443
Protein crystallography data
The structure of Structure of HDM2 with Dimer Inducing Indolyl Hydantoin Ro-2443, PDB code: 3vbg
was solved by
C.M.Lukacs,
C.A.Janson,
B.J.Graves,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 30.00 / 2.80 |
| Space group | P 32 |
| Cell size a, b, c (Å), α, β, γ (°) | 64.392, 64.392, 167.587, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 18.2 / 22 |
Other elements in 3vbg:
The structure of Structure of HDM2 with Dimer Inducing Indolyl Hydantoin Ro-2443 also contains other interesting chemical elements:
| Fluorine | (F) | 8 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Structure of HDM2 with Dimer Inducing Indolyl Hydantoin Ro-2443
(pdb code 3vbg). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 4 binding sites of Chlorine where determined in the Structure of HDM2 with Dimer Inducing Indolyl Hydantoin Ro-2443, PDB code: 3vbg:
Jump to Chlorine binding site number: 1; 2; 3; 4;
In total 4 binding sites of Chlorine where determined in the Structure of HDM2 with Dimer Inducing Indolyl Hydantoin Ro-2443, PDB code: 3vbg:
Jump to Chlorine binding site number: 1; 2; 3; 4;
Chlorine binding site 1 out of 4 in 3vbg
Go back to
Chlorine binding site 1 out
of 4 in the Structure of HDM2 with Dimer Inducing Indolyl Hydantoin Ro-2443
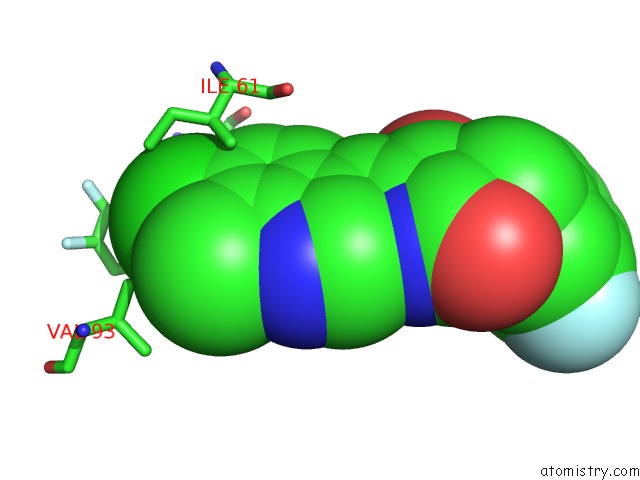
Mono view
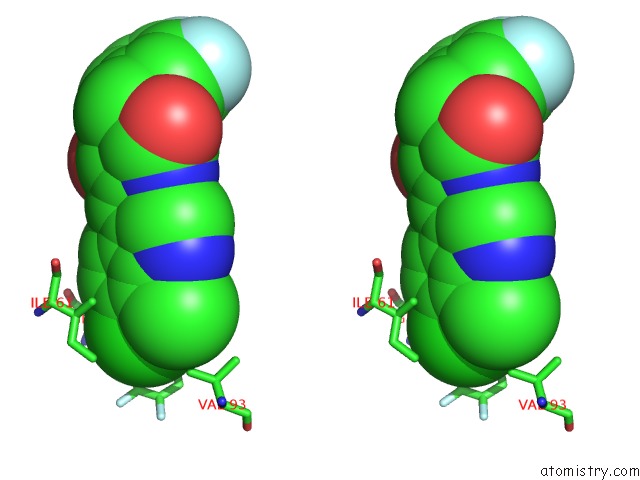
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Structure of HDM2 with Dimer Inducing Indolyl Hydantoin Ro-2443 within 5.0Å range:
|
Chlorine binding site 2 out of 4 in 3vbg
Go back to
Chlorine binding site 2 out
of 4 in the Structure of HDM2 with Dimer Inducing Indolyl Hydantoin Ro-2443
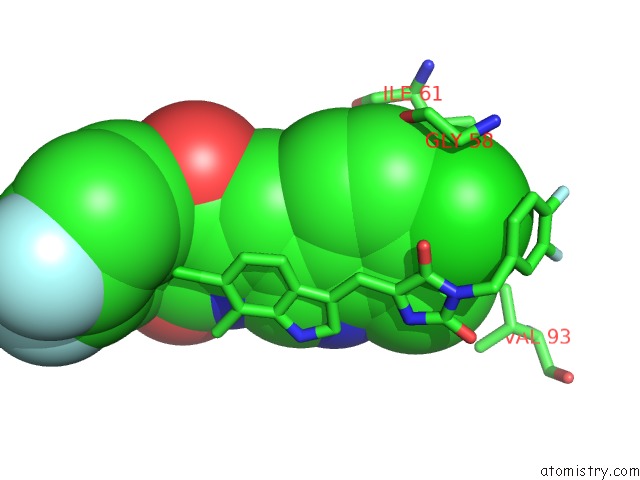
Mono view
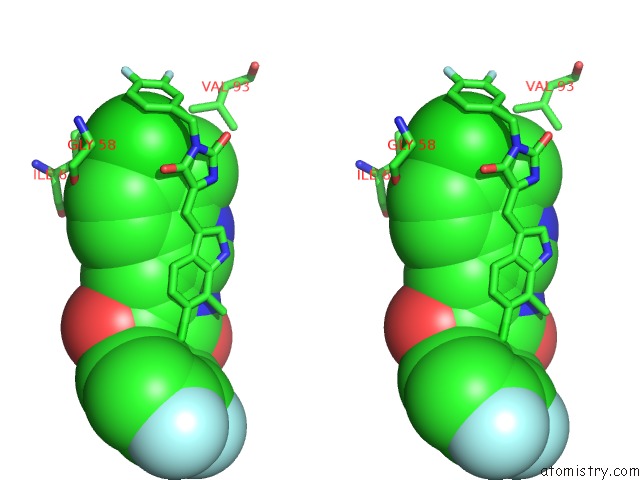
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Structure of HDM2 with Dimer Inducing Indolyl Hydantoin Ro-2443 within 5.0Å range:
|
Chlorine binding site 3 out of 4 in 3vbg
Go back to
Chlorine binding site 3 out
of 4 in the Structure of HDM2 with Dimer Inducing Indolyl Hydantoin Ro-2443
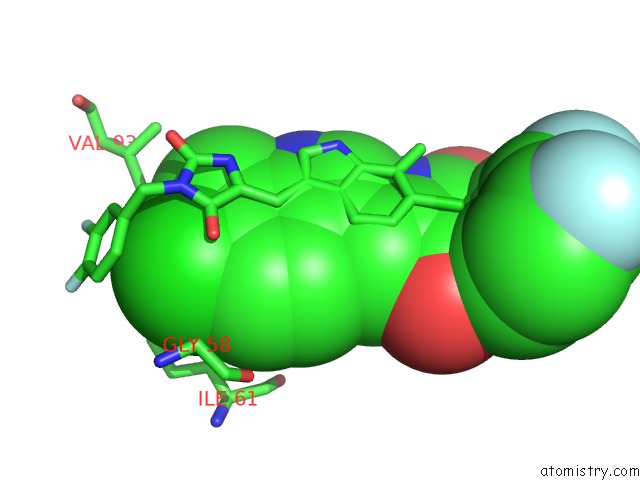
Mono view
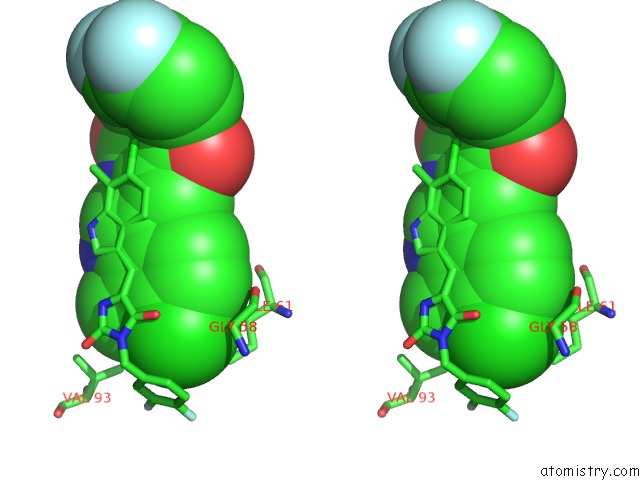
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Structure of HDM2 with Dimer Inducing Indolyl Hydantoin Ro-2443 within 5.0Å range:
|
Chlorine binding site 4 out of 4 in 3vbg
Go back to
Chlorine binding site 4 out
of 4 in the Structure of HDM2 with Dimer Inducing Indolyl Hydantoin Ro-2443
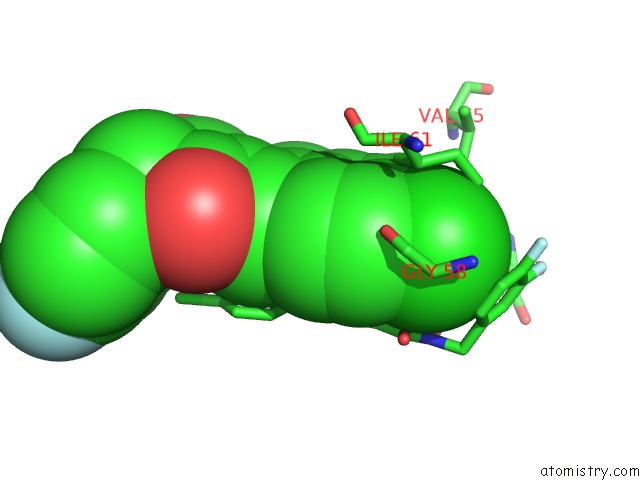
Mono view
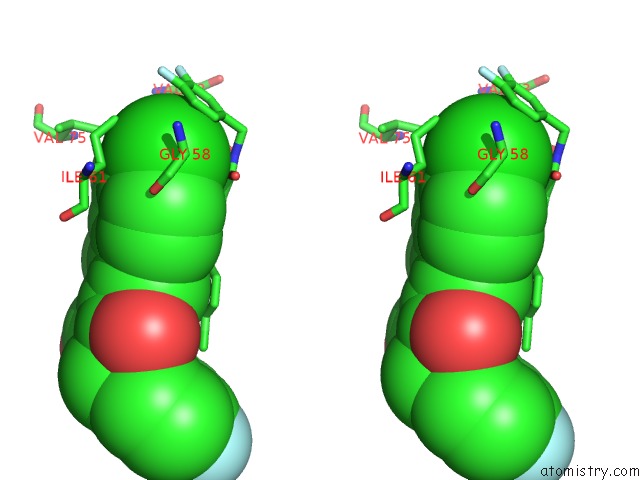
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Structure of HDM2 with Dimer Inducing Indolyl Hydantoin Ro-2443 within 5.0Å range:
|
Reference:
B.Graves,
T.Thompson,
M.Xia,
C.Janson,
C.Lukacs,
D.Deo,
P.Di Lello,
D.Fry,
C.Garvie,
K.S.Huang,
L.Gao,
C.Tovar,
A.Lovey,
J.Wanner,
L.T.Vassilev.
Activation of the P53 Pathway By Small-Molecule-Induced MDM2 and Mdmx Dimerization. Proc.Natl.Acad.Sci.Usa V. 109 11788 2012.
ISSN: ISSN 0027-8424
PubMed: 22745160
DOI: 10.1073/PNAS.1203789109
Page generated: Sun Jul 21 06:48:11 2024
ISSN: ISSN 0027-8424
PubMed: 22745160
DOI: 10.1073/PNAS.1203789109
Last articles
Ca in 5NGYCa in 5NG1
Ca in 5NFZ
Ca in 5NF3
Ca in 5NF2
Ca in 5NF0
Ca in 5NEY
Ca in 5NES
Ca in 5NEU
Ca in 5NET