Chlorine »
PDB 3wns-3x1v »
3wtm »
Chlorine in PDB 3wtm: Crystal Structure of Lymnaea Stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed with Nitromethylene Analogue of Imidacloprid
Protein crystallography data
The structure of Crystal Structure of Lymnaea Stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed with Nitromethylene Analogue of Imidacloprid, PDB code: 3wtm
was solved by
T.Okajima,
M.Ihara,
A.Yamashita,
T.Oda,
K.Matsuda,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 17.97 / 2.48 |
| Space group | P 65 |
| Cell size a, b, c (Å), α, β, γ (°) | 74.812, 74.812, 351.402, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 20.3 / 26.2 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of Lymnaea Stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed with Nitromethylene Analogue of Imidacloprid
(pdb code 3wtm). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 5 binding sites of Chlorine where determined in the Crystal Structure of Lymnaea Stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed with Nitromethylene Analogue of Imidacloprid, PDB code: 3wtm:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5;
In total 5 binding sites of Chlorine where determined in the Crystal Structure of Lymnaea Stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed with Nitromethylene Analogue of Imidacloprid, PDB code: 3wtm:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5;
Chlorine binding site 1 out of 5 in 3wtm
Go back to
Chlorine binding site 1 out
of 5 in the Crystal Structure of Lymnaea Stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed with Nitromethylene Analogue of Imidacloprid
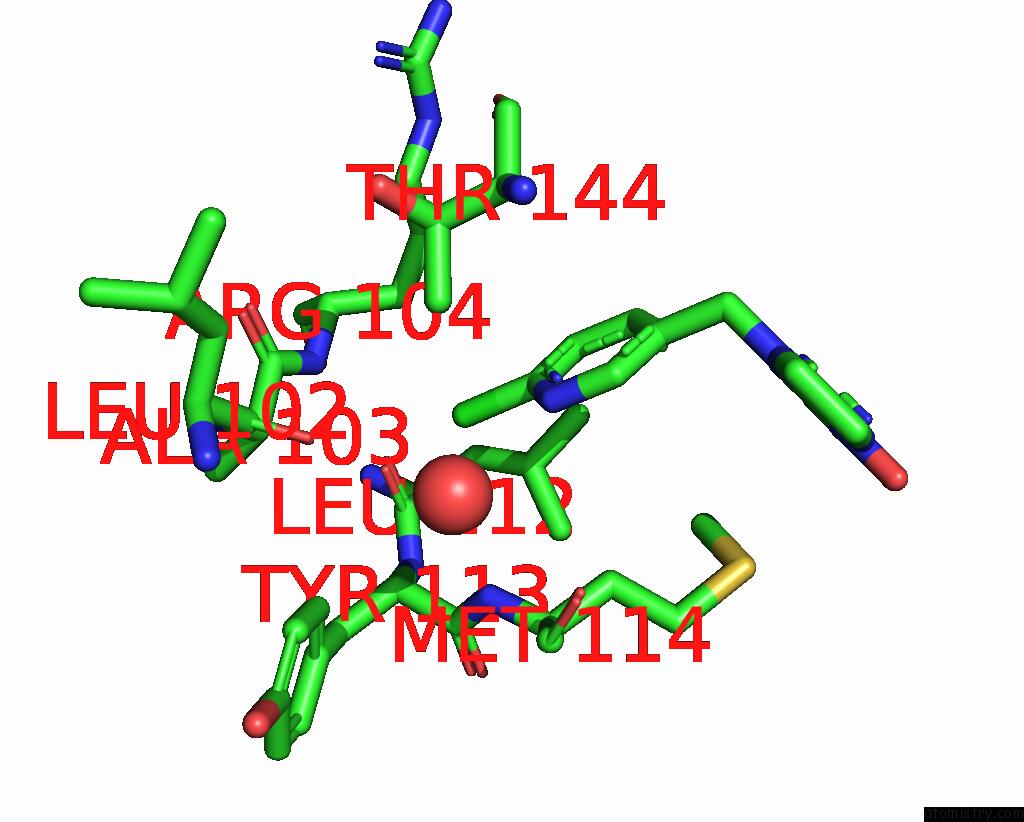
Mono view
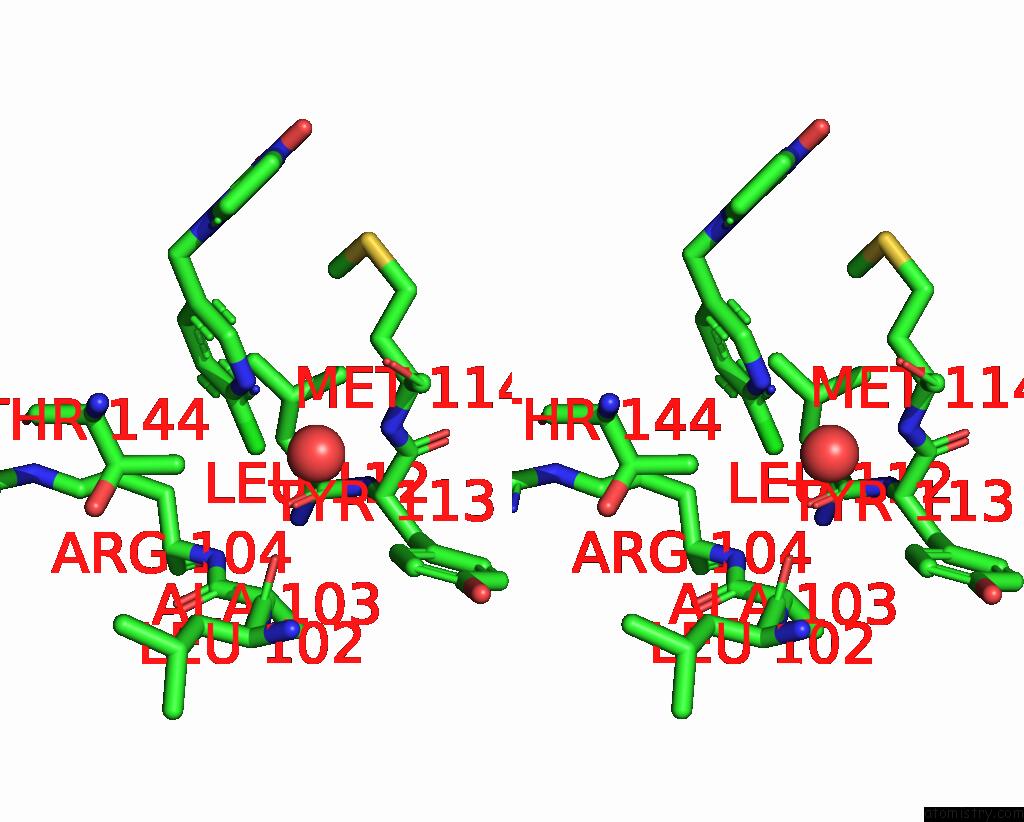
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of Lymnaea Stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed with Nitromethylene Analogue of Imidacloprid within 5.0Å range:
|
Chlorine binding site 2 out of 5 in 3wtm
Go back to
Chlorine binding site 2 out
of 5 in the Crystal Structure of Lymnaea Stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed with Nitromethylene Analogue of Imidacloprid
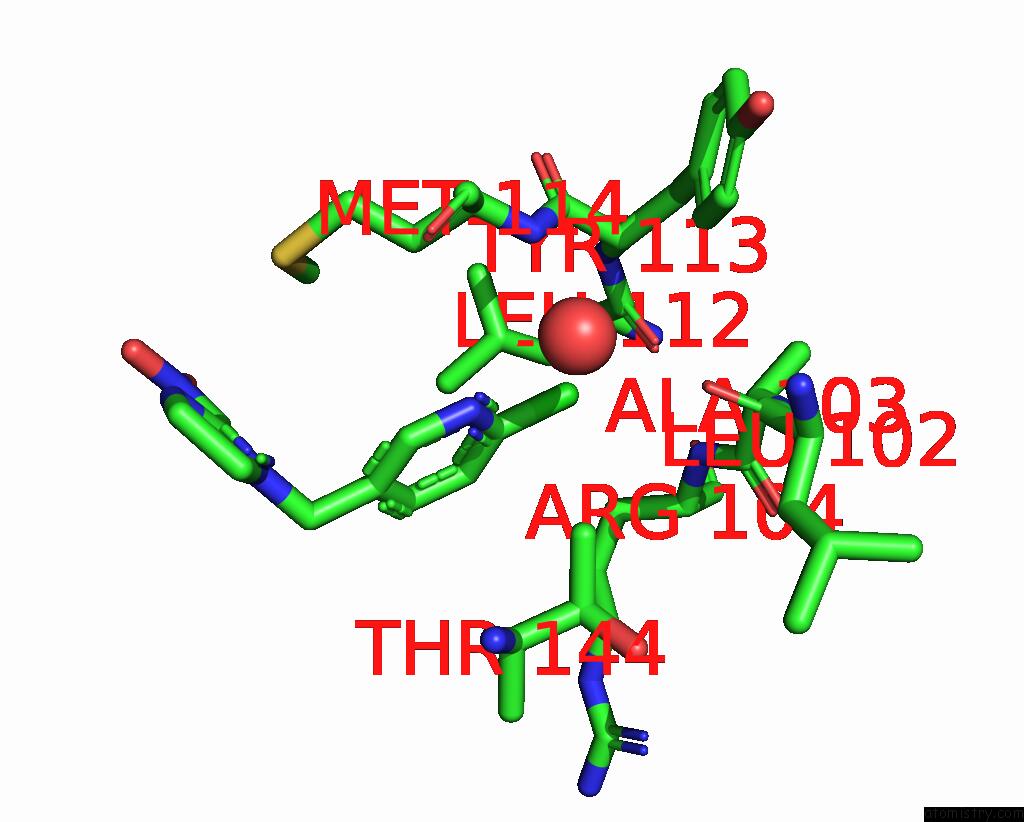
Mono view
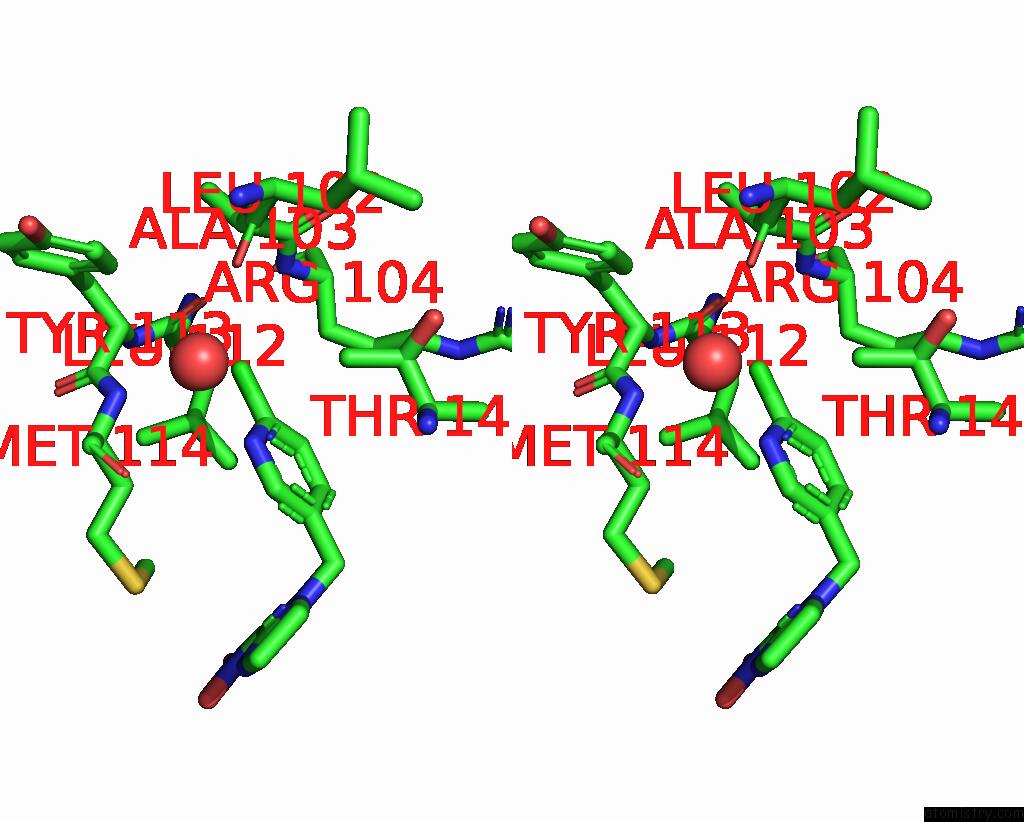
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of Lymnaea Stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed with Nitromethylene Analogue of Imidacloprid within 5.0Å range:
|
Chlorine binding site 3 out of 5 in 3wtm
Go back to
Chlorine binding site 3 out
of 5 in the Crystal Structure of Lymnaea Stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed with Nitromethylene Analogue of Imidacloprid
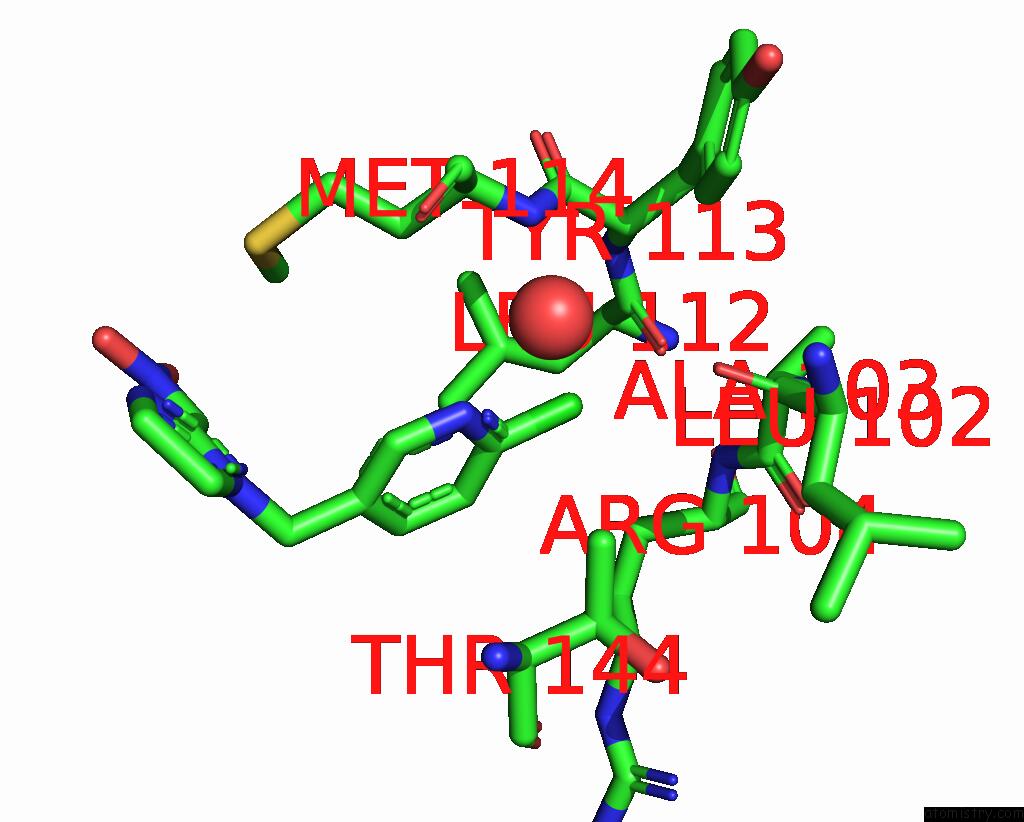
Mono view
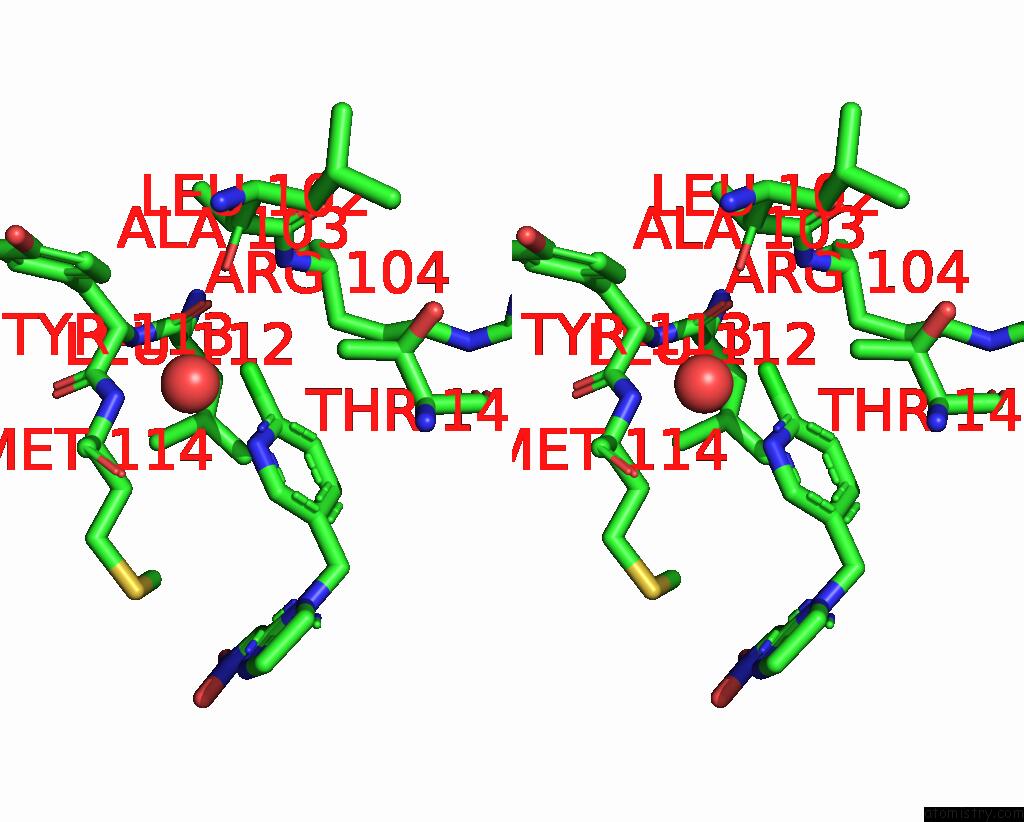
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Crystal Structure of Lymnaea Stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed with Nitromethylene Analogue of Imidacloprid within 5.0Å range:
|
Chlorine binding site 4 out of 5 in 3wtm
Go back to
Chlorine binding site 4 out
of 5 in the Crystal Structure of Lymnaea Stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed with Nitromethylene Analogue of Imidacloprid
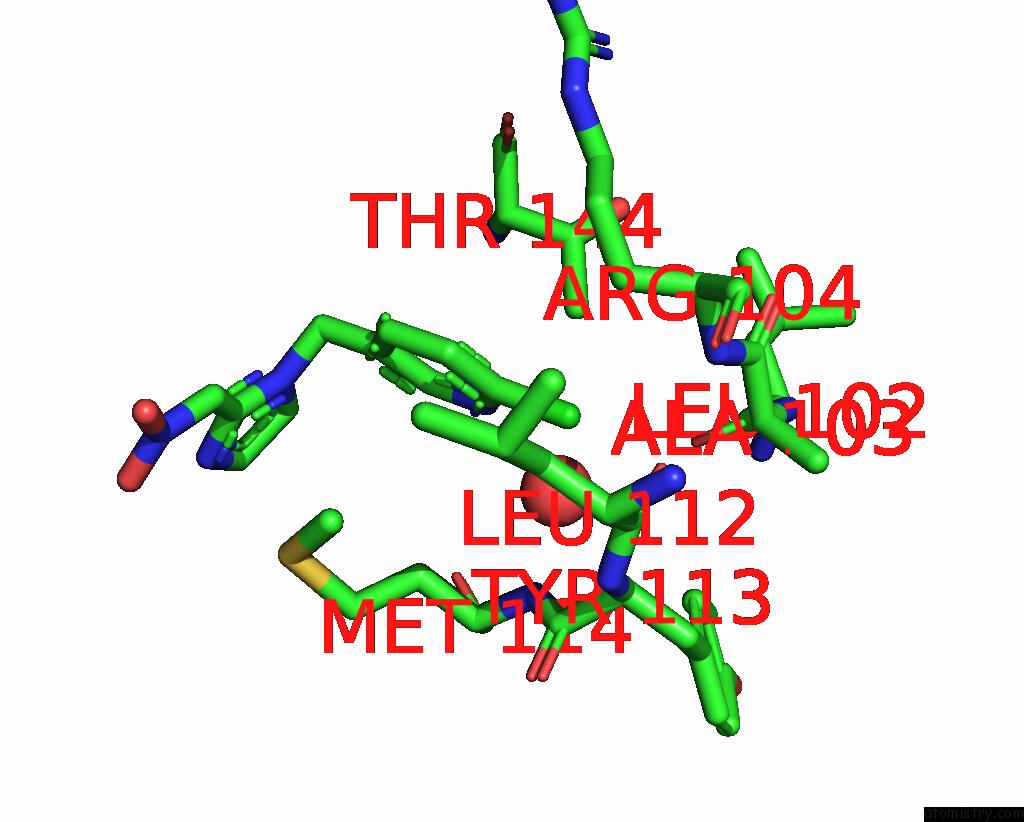
Mono view
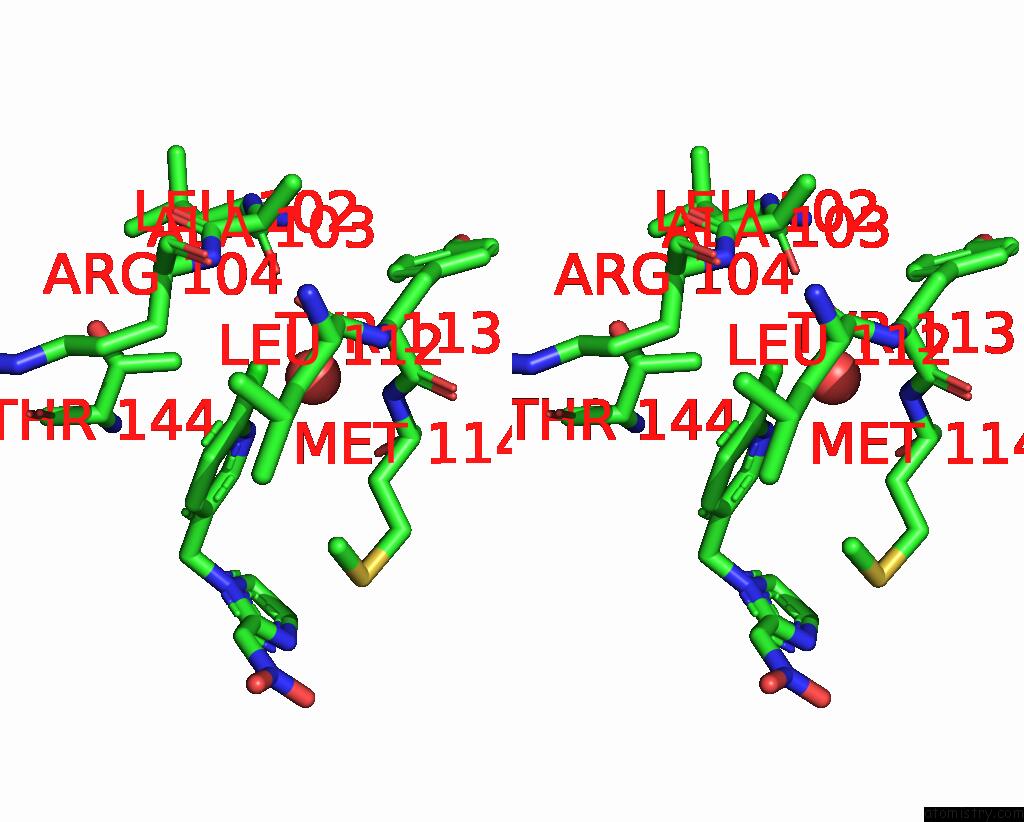
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Crystal Structure of Lymnaea Stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed with Nitromethylene Analogue of Imidacloprid within 5.0Å range:
|
Chlorine binding site 5 out of 5 in 3wtm
Go back to
Chlorine binding site 5 out
of 5 in the Crystal Structure of Lymnaea Stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed with Nitromethylene Analogue of Imidacloprid
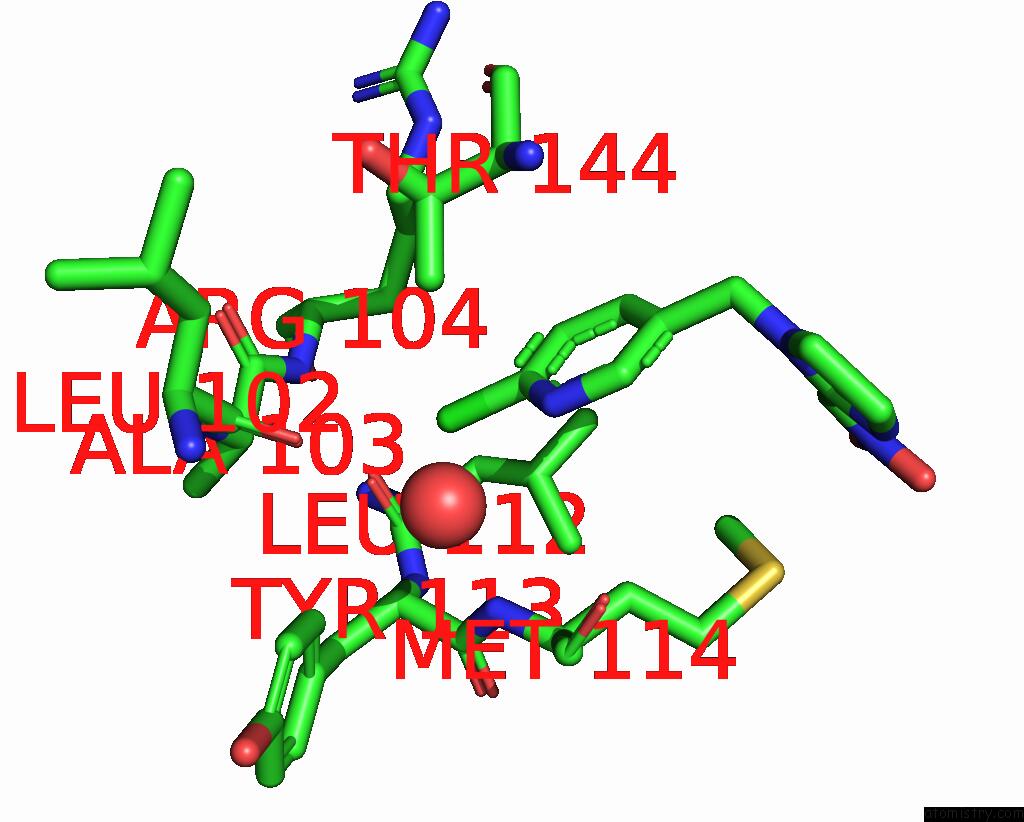
Mono view
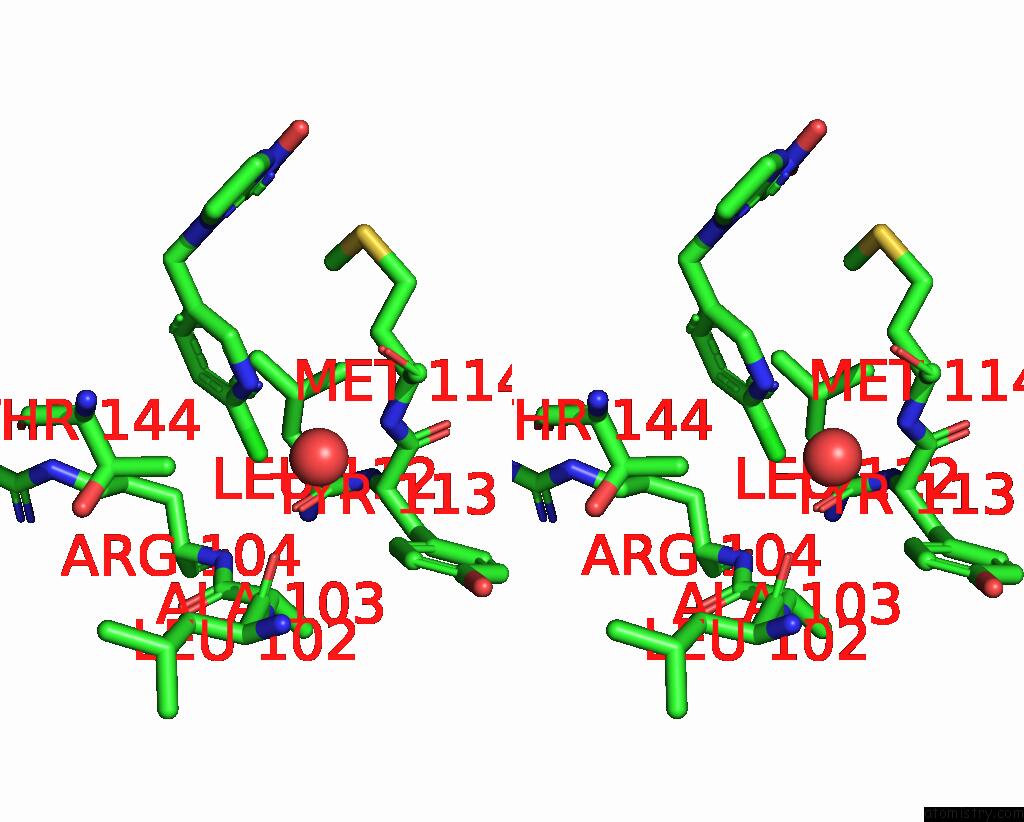
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Crystal Structure of Lymnaea Stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed with Nitromethylene Analogue of Imidacloprid within 5.0Å range:
|
Reference:
M.Ihara,
T.Okajima,
A.Yamashita,
T.Oda,
T.Asano,
M.Matsui,
D.B.Sattelle,
K.Matsuda.
Studies on An Acetylcholine Binding Protein Identify A Basic Residue in Loop G on the Beta 1 Strand As A New Structural Determinant of Neonicotinoid Actions Mol.Pharmacol. V. 86 736 2014.
ISSN: ISSN 0026-895X
PubMed: 25267717
DOI: 10.1124/MOL.114.094698
Page generated: Fri Jul 11 12:05:51 2025
ISSN: ISSN 0026-895X
PubMed: 25267717
DOI: 10.1124/MOL.114.094698
Last articles
Cl in 4LAUCl in 4LAG
Cl in 4LAB
Cl in 4L9Y
Cl in 4L9P
Cl in 4L3C
Cl in 4L29
Cl in 4L9O
Cl in 4L9D
Cl in 4L9I