Chlorine »
PDB 4dq9-4e02 »
4dtb »
Chlorine in PDB 4dtb: Crystal Structure of F95Y Aminoglycoside-2''-Phosphotransferase Type Iva in Complex with Guanosine
Protein crystallography data
The structure of Crystal Structure of F95Y Aminoglycoside-2''-Phosphotransferase Type Iva in Complex with Guanosine, PDB code: 4dtb
was solved by
K.Shi,
A.M.Berghuis,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 30.00 / 2.10 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 42.272, 101.341, 73.600, 90.00, 100.60, 90.00 |
| R / Rfree (%) | 19.7 / 25 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of F95Y Aminoglycoside-2''-Phosphotransferase Type Iva in Complex with Guanosine
(pdb code 4dtb). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the Crystal Structure of F95Y Aminoglycoside-2''-Phosphotransferase Type Iva in Complex with Guanosine, PDB code: 4dtb:
In total only one binding site of Chlorine was determined in the Crystal Structure of F95Y Aminoglycoside-2''-Phosphotransferase Type Iva in Complex with Guanosine, PDB code: 4dtb:
Chlorine binding site 1 out of 1 in 4dtb
Go back to
Chlorine binding site 1 out
of 1 in the Crystal Structure of F95Y Aminoglycoside-2''-Phosphotransferase Type Iva in Complex with Guanosine
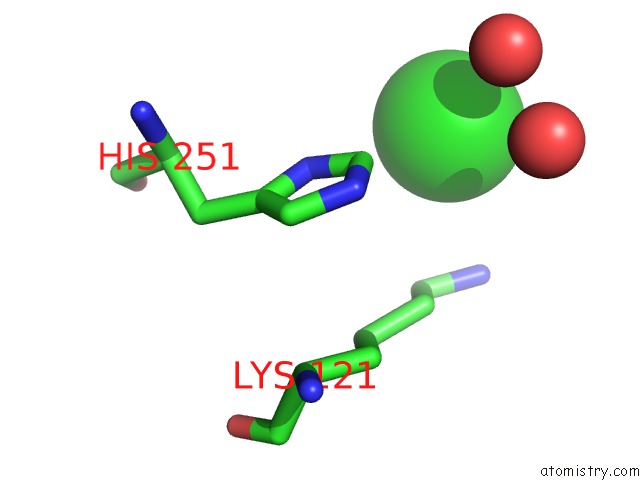
Mono view
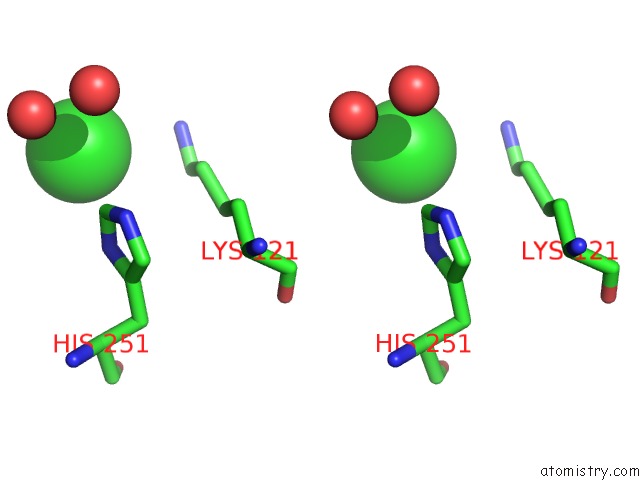
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of F95Y Aminoglycoside-2''-Phosphotransferase Type Iva in Complex with Guanosine within 5.0Å range:
|
Reference:
K.Shi,
A.M.Berghuis.
Structural Basis For Dual Nucleotide Selectivity of Aminoglycoside 2''-Phosphotransferase Iva Provides Insight on Determinants of Nucleotide Specificity of Aminoglycoside Kinases. J.Biol.Chem. V. 287 13094 2012.
ISSN: ISSN 0021-9258
PubMed: 22371504
DOI: 10.1074/JBC.M112.349670
Page generated: Fri Jul 11 14:31:12 2025
ISSN: ISSN 0021-9258
PubMed: 22371504
DOI: 10.1074/JBC.M112.349670
Last articles
Cl in 4UGJCl in 4UGI
Cl in 4UGH
Cl in 4UGG
Cl in 4UGE
Cl in 4UGF
Cl in 4UGD
Cl in 4UGC
Cl in 4UG9
Cl in 4UGA