Chlorine »
PDB 4fmg-4fv2 »
4fsl »
Chlorine in PDB 4fsl: Crystal Structure of Beta-Site App-Cleaving Enzyme 1 (Bace-Db-Mut) Complex with N-(N-(4- Acetamido-3-Chloro-5-Methylbenzyl) Carbamimidoyl)-3-(4- Methoxyphenyl)-5-Methyl-4-Isothiazolecarboxamide
Enzymatic activity of Crystal Structure of Beta-Site App-Cleaving Enzyme 1 (Bace-Db-Mut) Complex with N-(N-(4- Acetamido-3-Chloro-5-Methylbenzyl) Carbamimidoyl)-3-(4- Methoxyphenyl)-5-Methyl-4-Isothiazolecarboxamide
All present enzymatic activity of Crystal Structure of Beta-Site App-Cleaving Enzyme 1 (Bace-Db-Mut) Complex with N-(N-(4- Acetamido-3-Chloro-5-Methylbenzyl) Carbamimidoyl)-3-(4- Methoxyphenyl)-5-Methyl-4-Isothiazolecarboxamide:
3.4.23.46;
3.4.23.46;
Protein crystallography data
The structure of Crystal Structure of Beta-Site App-Cleaving Enzyme 1 (Bace-Db-Mut) Complex with N-(N-(4- Acetamido-3-Chloro-5-Methylbenzyl) Carbamimidoyl)-3-(4- Methoxyphenyl)-5-Methyl-4-Isothiazolecarboxamide, PDB code: 4fsl
was solved by
J.K.Muckelbauer,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 50.00 / 2.50 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 86.540, 131.177, 90.315, 90.00, 97.61, 90.00 |
| R / Rfree (%) | 22.4 / 27.7 |
Other elements in 4fsl:
The structure of Crystal Structure of Beta-Site App-Cleaving Enzyme 1 (Bace-Db-Mut) Complex with N-(N-(4- Acetamido-3-Chloro-5-Methylbenzyl) Carbamimidoyl)-3-(4- Methoxyphenyl)-5-Methyl-4-Isothiazolecarboxamide also contains other interesting chemical elements:
| Iodine | (I) | 12 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of Beta-Site App-Cleaving Enzyme 1 (Bace-Db-Mut) Complex with N-(N-(4- Acetamido-3-Chloro-5-Methylbenzyl) Carbamimidoyl)-3-(4- Methoxyphenyl)-5-Methyl-4-Isothiazolecarboxamide
(pdb code 4fsl). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 4 binding sites of Chlorine where determined in the Crystal Structure of Beta-Site App-Cleaving Enzyme 1 (Bace-Db-Mut) Complex with N-(N-(4- Acetamido-3-Chloro-5-Methylbenzyl) Carbamimidoyl)-3-(4- Methoxyphenyl)-5-Methyl-4-Isothiazolecarboxamide, PDB code: 4fsl:
Jump to Chlorine binding site number: 1; 2; 3; 4;
In total 4 binding sites of Chlorine where determined in the Crystal Structure of Beta-Site App-Cleaving Enzyme 1 (Bace-Db-Mut) Complex with N-(N-(4- Acetamido-3-Chloro-5-Methylbenzyl) Carbamimidoyl)-3-(4- Methoxyphenyl)-5-Methyl-4-Isothiazolecarboxamide, PDB code: 4fsl:
Jump to Chlorine binding site number: 1; 2; 3; 4;
Chlorine binding site 1 out of 4 in 4fsl
Go back to
Chlorine binding site 1 out
of 4 in the Crystal Structure of Beta-Site App-Cleaving Enzyme 1 (Bace-Db-Mut) Complex with N-(N-(4- Acetamido-3-Chloro-5-Methylbenzyl) Carbamimidoyl)-3-(4- Methoxyphenyl)-5-Methyl-4-Isothiazolecarboxamide
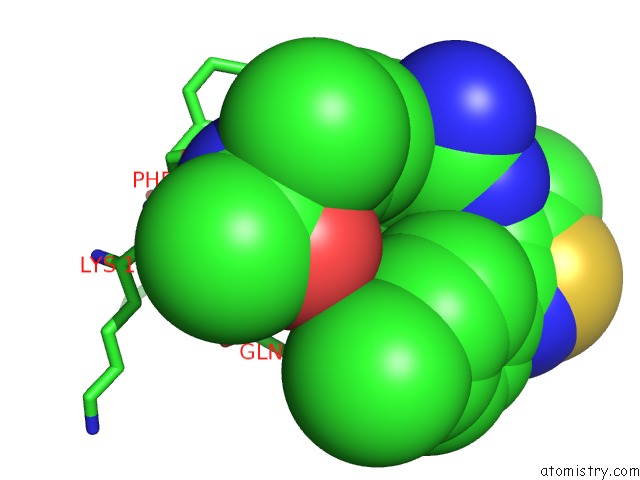
Mono view
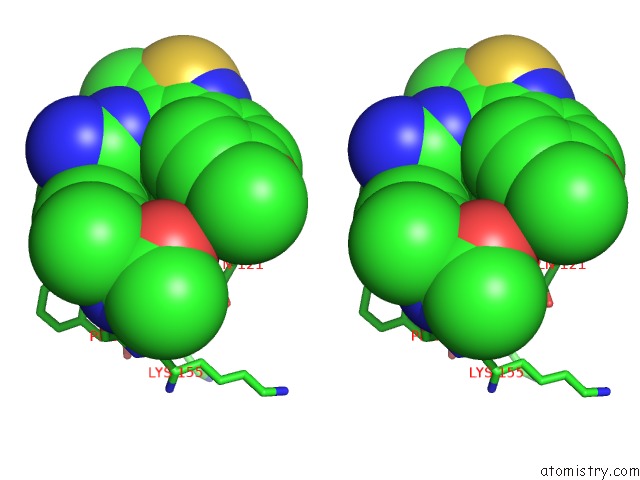
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of Beta-Site App-Cleaving Enzyme 1 (Bace-Db-Mut) Complex with N-(N-(4- Acetamido-3-Chloro-5-Methylbenzyl) Carbamimidoyl)-3-(4- Methoxyphenyl)-5-Methyl-4-Isothiazolecarboxamide within 5.0Å range:
|
Chlorine binding site 2 out of 4 in 4fsl
Go back to
Chlorine binding site 2 out
of 4 in the Crystal Structure of Beta-Site App-Cleaving Enzyme 1 (Bace-Db-Mut) Complex with N-(N-(4- Acetamido-3-Chloro-5-Methylbenzyl) Carbamimidoyl)-3-(4- Methoxyphenyl)-5-Methyl-4-Isothiazolecarboxamide
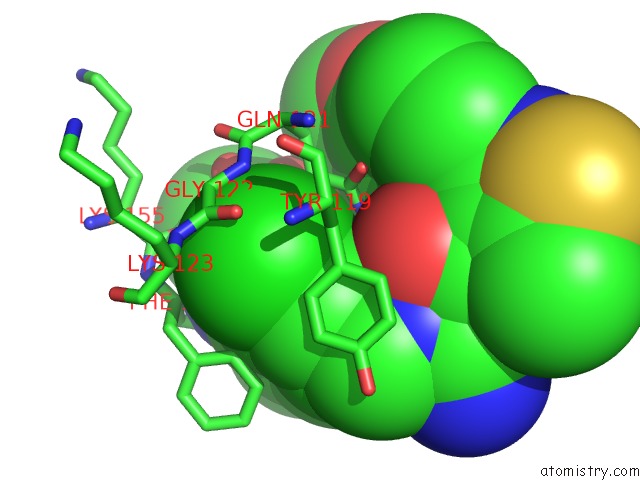
Mono view
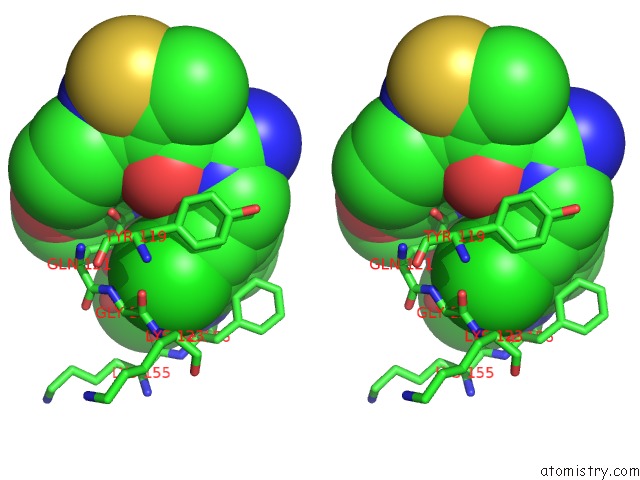
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of Beta-Site App-Cleaving Enzyme 1 (Bace-Db-Mut) Complex with N-(N-(4- Acetamido-3-Chloro-5-Methylbenzyl) Carbamimidoyl)-3-(4- Methoxyphenyl)-5-Methyl-4-Isothiazolecarboxamide within 5.0Å range:
|
Chlorine binding site 3 out of 4 in 4fsl
Go back to
Chlorine binding site 3 out
of 4 in the Crystal Structure of Beta-Site App-Cleaving Enzyme 1 (Bace-Db-Mut) Complex with N-(N-(4- Acetamido-3-Chloro-5-Methylbenzyl) Carbamimidoyl)-3-(4- Methoxyphenyl)-5-Methyl-4-Isothiazolecarboxamide
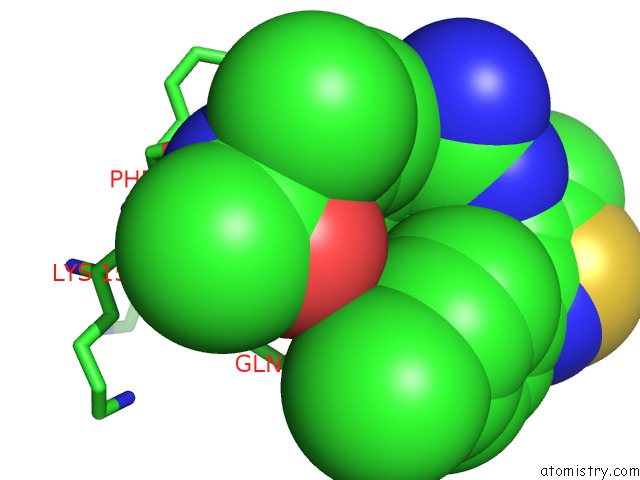
Mono view
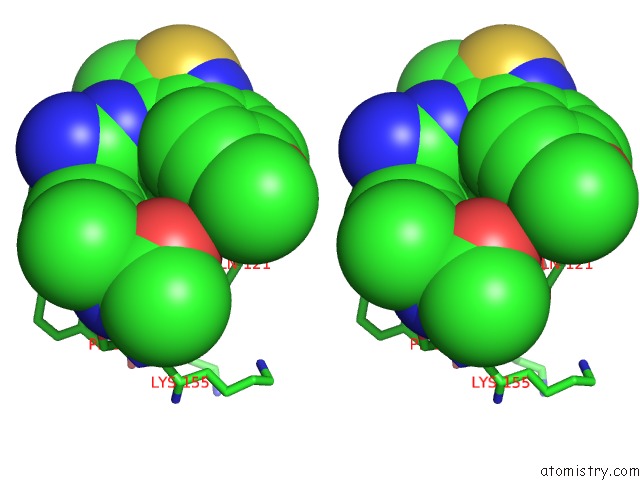
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Crystal Structure of Beta-Site App-Cleaving Enzyme 1 (Bace-Db-Mut) Complex with N-(N-(4- Acetamido-3-Chloro-5-Methylbenzyl) Carbamimidoyl)-3-(4- Methoxyphenyl)-5-Methyl-4-Isothiazolecarboxamide within 5.0Å range:
|
Chlorine binding site 4 out of 4 in 4fsl
Go back to
Chlorine binding site 4 out
of 4 in the Crystal Structure of Beta-Site App-Cleaving Enzyme 1 (Bace-Db-Mut) Complex with N-(N-(4- Acetamido-3-Chloro-5-Methylbenzyl) Carbamimidoyl)-3-(4- Methoxyphenyl)-5-Methyl-4-Isothiazolecarboxamide
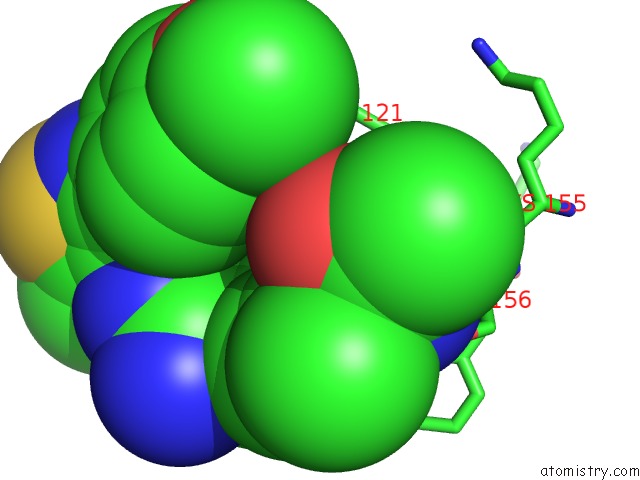
Mono view
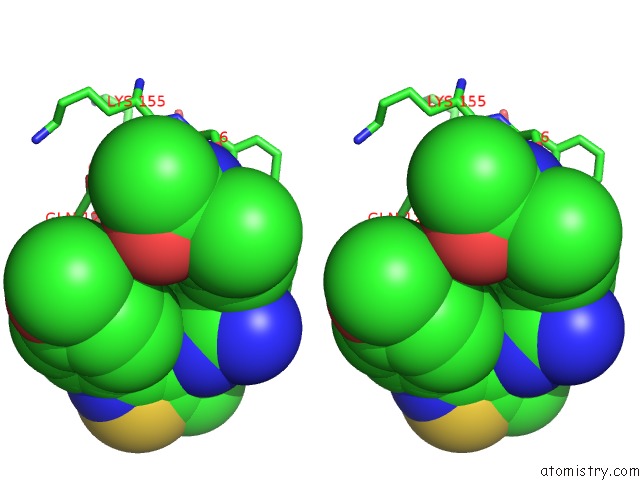
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Crystal Structure of Beta-Site App-Cleaving Enzyme 1 (Bace-Db-Mut) Complex with N-(N-(4- Acetamido-3-Chloro-5-Methylbenzyl) Carbamimidoyl)-3-(4- Methoxyphenyl)-5-Methyl-4-Isothiazolecarboxamide within 5.0Å range:
|
Reference:
S.W.Gerritz,
W.Zhai,
S.Shi,
S.Zhu,
J.H.Toyn,
J.E.Meredith,
L.G.Iben,
C.R.Burton,
C.F.Albright,
A.C.Good,
A.J.Tebben,
J.K.Muckelbauer,
D.M.Camac,
W.Metzler,
L.S.Cook,
R.Padmanabha,
K.A.Lentz,
M.J.Sofia,
M.A.Poss,
J.E.Macor,
L.A.Thompson.
Acyl Guanidine Inhibitors of Beta-Secretase (Bace-1): Optimization of A Micromolar Hit to A Nanomolar Lead Via Iterative Solid- and Solution-Phase Library Synthesis J.Med.Chem. V. 55 9208 2012.
ISSN: ISSN 0022-2623
PubMed: 23030502
DOI: 10.1021/JM300931Y
Page generated: Fri Jul 11 15:22:51 2025
ISSN: ISSN 0022-2623
PubMed: 23030502
DOI: 10.1021/JM300931Y
Last articles
F in 4JYWF in 4K0Y
F in 4JZ1
F in 4JZ0
F in 4JX7
F in 4JVS
F in 4JVR
F in 4JU1
F in 4JUO
F in 4JVE