Chlorine »
PDB 4fv3-4g7y »
4fwb »
Chlorine in PDB 4fwb: Structure of Rhodococcus Rhodochrous Haloalkane Dehalogenase Mutant DHAA31 in Complex with 1, 2, 3 - Trichloropropane
Enzymatic activity of Structure of Rhodococcus Rhodochrous Haloalkane Dehalogenase Mutant DHAA31 in Complex with 1, 2, 3 - Trichloropropane
All present enzymatic activity of Structure of Rhodococcus Rhodochrous Haloalkane Dehalogenase Mutant DHAA31 in Complex with 1, 2, 3 - Trichloropropane:
3.8.1.5;
3.8.1.5;
Protein crystallography data
The structure of Structure of Rhodococcus Rhodochrous Haloalkane Dehalogenase Mutant DHAA31 in Complex with 1, 2, 3 - Trichloropropane, PDB code: 4fwb
was solved by
M.Lahoda,
A.Stsiapanava,
J.Mesters,
I.Kuta Smatanova,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 10.00 / 1.26 |
| Space group | P 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 42.498, 44.395, 46.538, 115.32, 97.70, 109.58 |
| R / Rfree (%) | 13.6 / 16.6 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Structure of Rhodococcus Rhodochrous Haloalkane Dehalogenase Mutant DHAA31 in Complex with 1, 2, 3 - Trichloropropane
(pdb code 4fwb). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 7 binding sites of Chlorine where determined in the Structure of Rhodococcus Rhodochrous Haloalkane Dehalogenase Mutant DHAA31 in Complex with 1, 2, 3 - Trichloropropane, PDB code: 4fwb:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7;
In total 7 binding sites of Chlorine where determined in the Structure of Rhodococcus Rhodochrous Haloalkane Dehalogenase Mutant DHAA31 in Complex with 1, 2, 3 - Trichloropropane, PDB code: 4fwb:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7;
Chlorine binding site 1 out of 7 in 4fwb
Go back to
Chlorine binding site 1 out
of 7 in the Structure of Rhodococcus Rhodochrous Haloalkane Dehalogenase Mutant DHAA31 in Complex with 1, 2, 3 - Trichloropropane
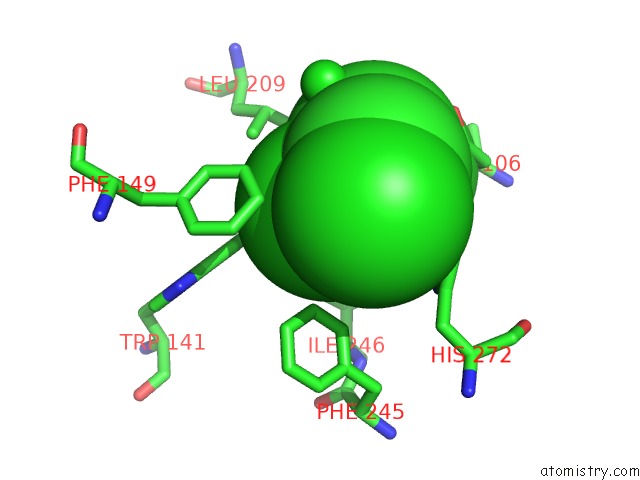
Mono view
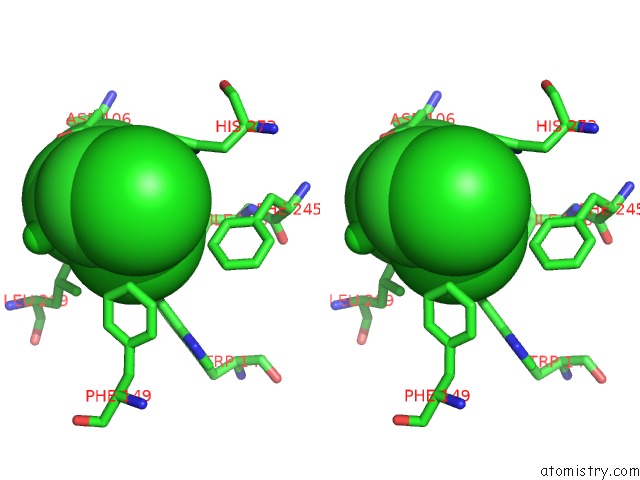
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Structure of Rhodococcus Rhodochrous Haloalkane Dehalogenase Mutant DHAA31 in Complex with 1, 2, 3 - Trichloropropane within 5.0Å range:
|
Chlorine binding site 2 out of 7 in 4fwb
Go back to
Chlorine binding site 2 out
of 7 in the Structure of Rhodococcus Rhodochrous Haloalkane Dehalogenase Mutant DHAA31 in Complex with 1, 2, 3 - Trichloropropane
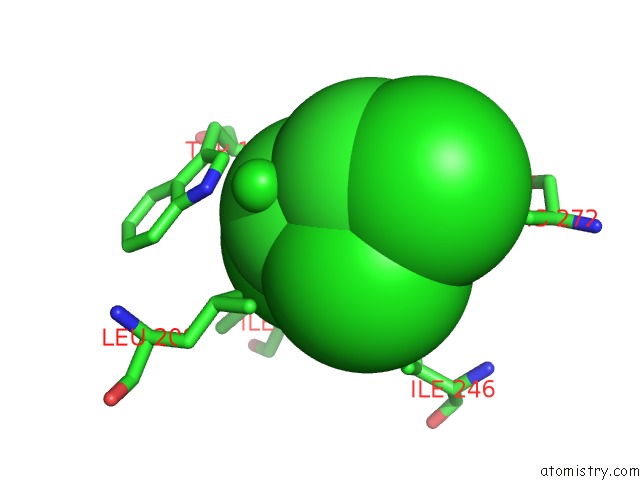
Mono view
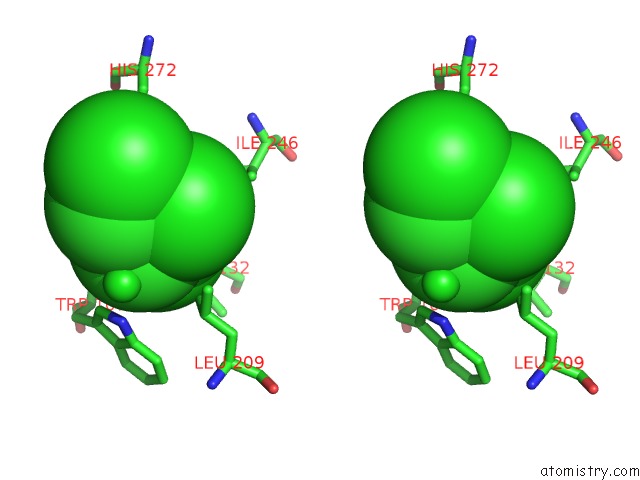
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Structure of Rhodococcus Rhodochrous Haloalkane Dehalogenase Mutant DHAA31 in Complex with 1, 2, 3 - Trichloropropane within 5.0Å range:
|
Chlorine binding site 3 out of 7 in 4fwb
Go back to
Chlorine binding site 3 out
of 7 in the Structure of Rhodococcus Rhodochrous Haloalkane Dehalogenase Mutant DHAA31 in Complex with 1, 2, 3 - Trichloropropane
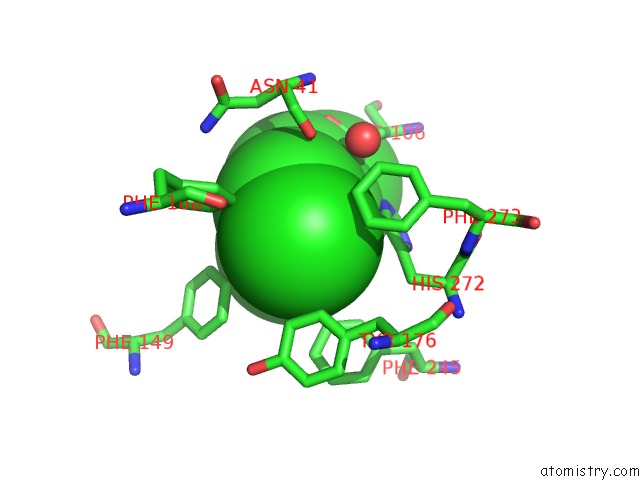
Mono view
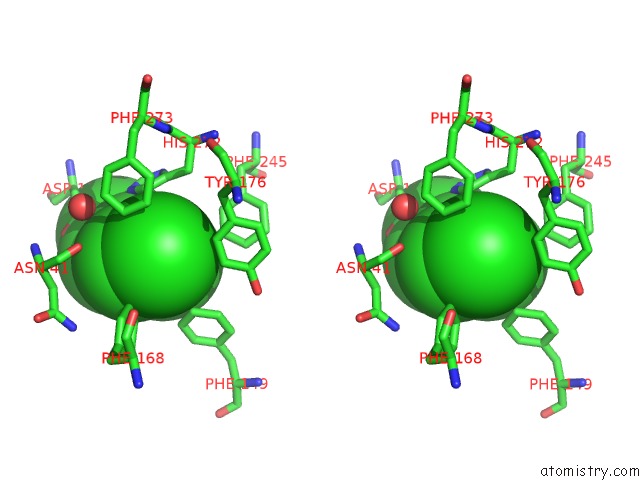
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Structure of Rhodococcus Rhodochrous Haloalkane Dehalogenase Mutant DHAA31 in Complex with 1, 2, 3 - Trichloropropane within 5.0Å range:
|
Chlorine binding site 4 out of 7 in 4fwb
Go back to
Chlorine binding site 4 out
of 7 in the Structure of Rhodococcus Rhodochrous Haloalkane Dehalogenase Mutant DHAA31 in Complex with 1, 2, 3 - Trichloropropane
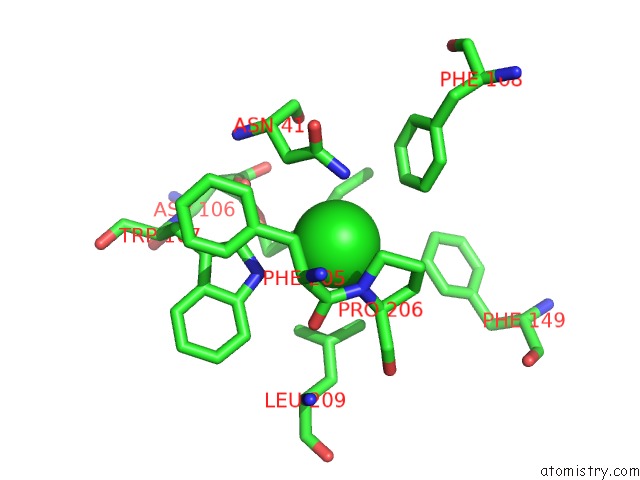
Mono view
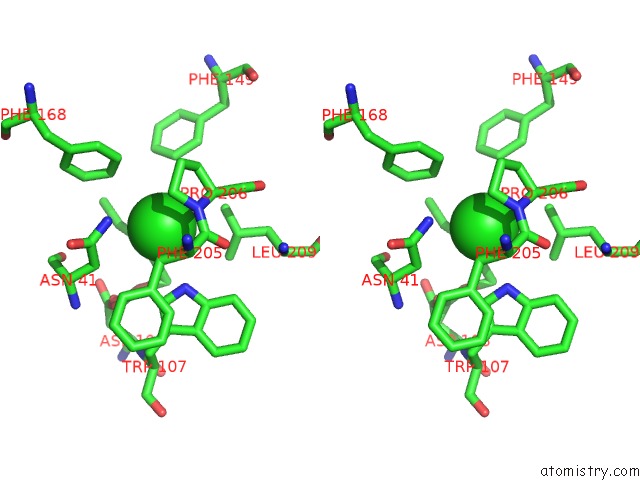
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Structure of Rhodococcus Rhodochrous Haloalkane Dehalogenase Mutant DHAA31 in Complex with 1, 2, 3 - Trichloropropane within 5.0Å range:
|
Chlorine binding site 5 out of 7 in 4fwb
Go back to
Chlorine binding site 5 out
of 7 in the Structure of Rhodococcus Rhodochrous Haloalkane Dehalogenase Mutant DHAA31 in Complex with 1, 2, 3 - Trichloropropane
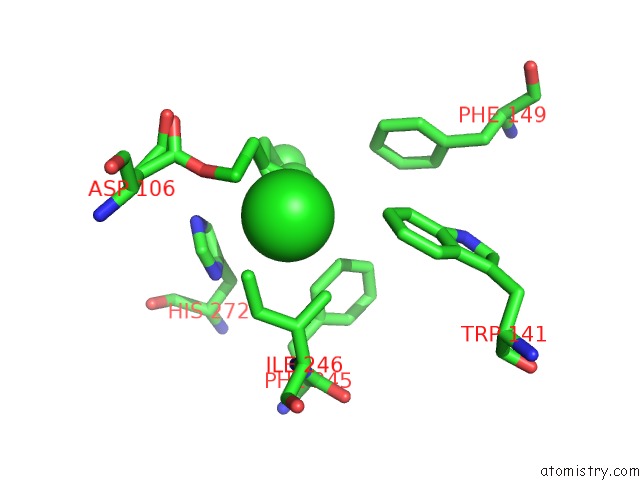
Mono view
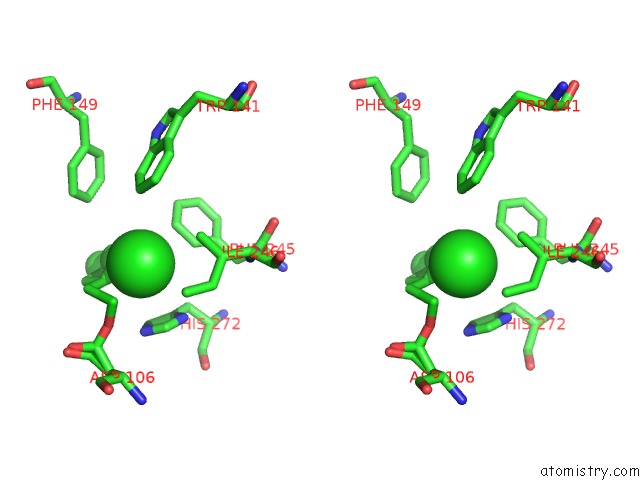
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Structure of Rhodococcus Rhodochrous Haloalkane Dehalogenase Mutant DHAA31 in Complex with 1, 2, 3 - Trichloropropane within 5.0Å range:
|
Chlorine binding site 6 out of 7 in 4fwb
Go back to
Chlorine binding site 6 out
of 7 in the Structure of Rhodococcus Rhodochrous Haloalkane Dehalogenase Mutant DHAA31 in Complex with 1, 2, 3 - Trichloropropane
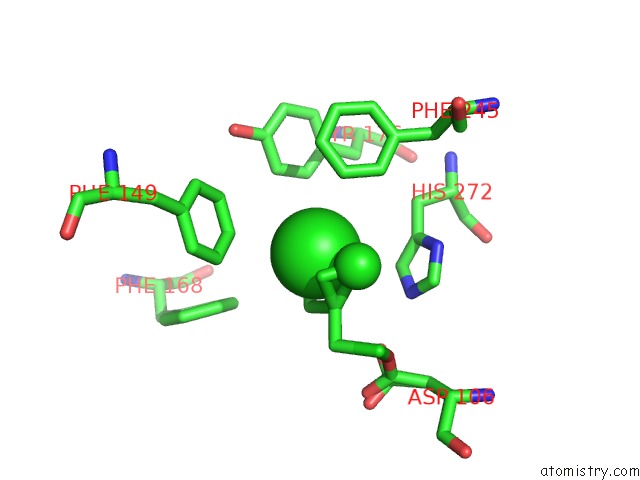
Mono view
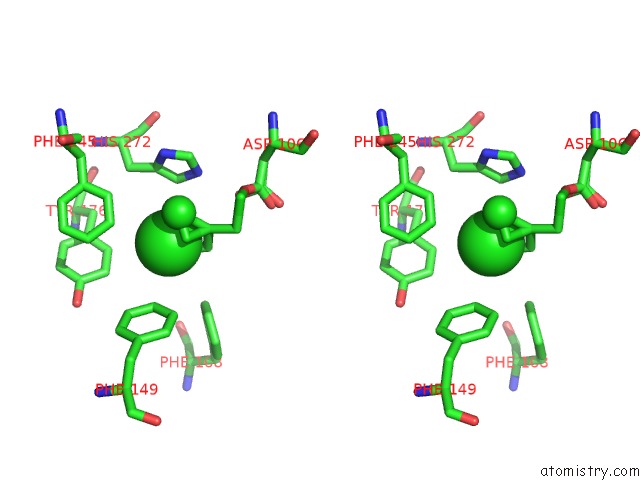
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 6 of Structure of Rhodococcus Rhodochrous Haloalkane Dehalogenase Mutant DHAA31 in Complex with 1, 2, 3 - Trichloropropane within 5.0Å range:
|
Chlorine binding site 7 out of 7 in 4fwb
Go back to
Chlorine binding site 7 out
of 7 in the Structure of Rhodococcus Rhodochrous Haloalkane Dehalogenase Mutant DHAA31 in Complex with 1, 2, 3 - Trichloropropane
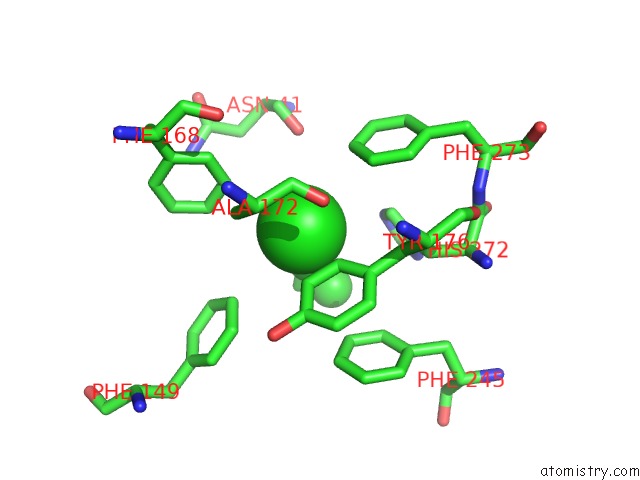
Mono view
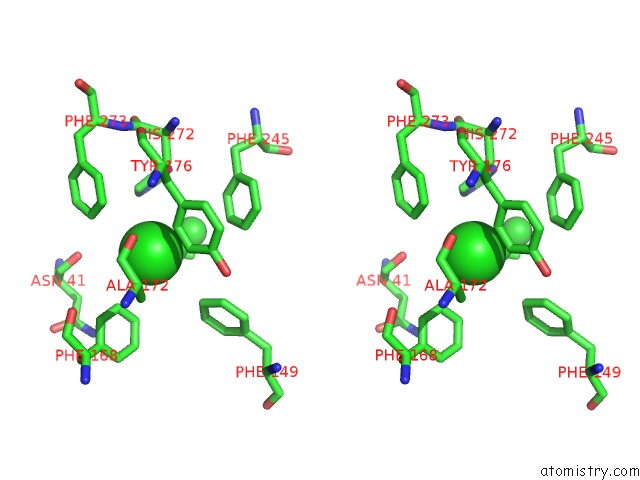
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 7 of Structure of Rhodococcus Rhodochrous Haloalkane Dehalogenase Mutant DHAA31 in Complex with 1, 2, 3 - Trichloropropane within 5.0Å range:
|
Reference:
M.Lahoda,
J.R.Mesters,
A.Stsiapanava,
R.Chaloupkova,
M.Kuty,
J.Damborsky,
I.Kuta Smatanova.
Crystallographic Analysis of 1,2,3-Trichloropropane Biodegradation By the Haloalkane Dehalogenase DHAA31. Acta Crystallogr.,Sect.D V. 70 209 2014.
ISSN: ISSN 0907-4449
PubMed: 24531456
DOI: 10.1107/S1399004713026254
Page generated: Fri Jul 11 15:25:54 2025
ISSN: ISSN 0907-4449
PubMed: 24531456
DOI: 10.1107/S1399004713026254
Last articles
Cl in 5VSBCl in 5VS7
Cl in 5VPB
Cl in 5VS0
Cl in 5VRZ
Cl in 5VRY
Cl in 5VQB
Cl in 5VRH
Cl in 5VRG
Cl in 5VPJ