Chlorine »
PDB 4he1-4hmr »
4hha »
Chlorine in PDB 4hha: Anti-Human Cytomegalovirus (Hcmv) Fab KE5 with Epitope Peptide Ad-2S1
Protein crystallography data
The structure of Anti-Human Cytomegalovirus (Hcmv) Fab KE5 with Epitope Peptide Ad-2S1, PDB code: 4hha
was solved by
S.Bryson,
L.Risnes,
S.Damgupta,
C.A.Thomson,
R.Pfoh,
J.W.Schrader,
E.F.Pai,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 43.00 / 1.60 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 43.250, 65.510, 67.110, 90.00, 96.58, 90.00 |
| R / Rfree (%) | 21.3 / 22.6 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Anti-Human Cytomegalovirus (Hcmv) Fab KE5 with Epitope Peptide Ad-2S1
(pdb code 4hha). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 5 binding sites of Chlorine where determined in the Anti-Human Cytomegalovirus (Hcmv) Fab KE5 with Epitope Peptide Ad-2S1, PDB code: 4hha:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5;
In total 5 binding sites of Chlorine where determined in the Anti-Human Cytomegalovirus (Hcmv) Fab KE5 with Epitope Peptide Ad-2S1, PDB code: 4hha:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5;
Chlorine binding site 1 out of 5 in 4hha
Go back to
Chlorine binding site 1 out
of 5 in the Anti-Human Cytomegalovirus (Hcmv) Fab KE5 with Epitope Peptide Ad-2S1
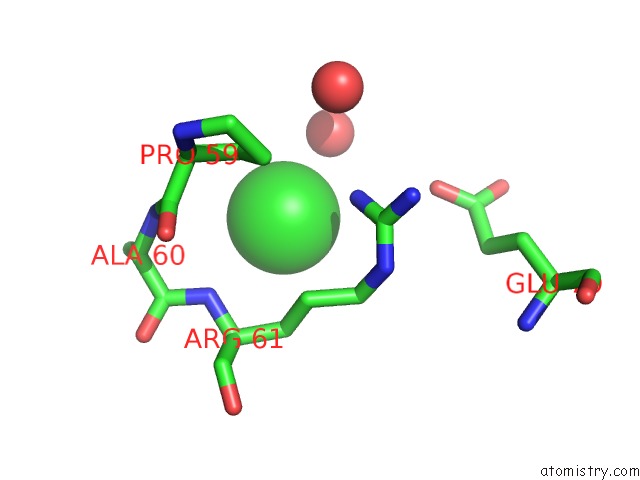
Mono view
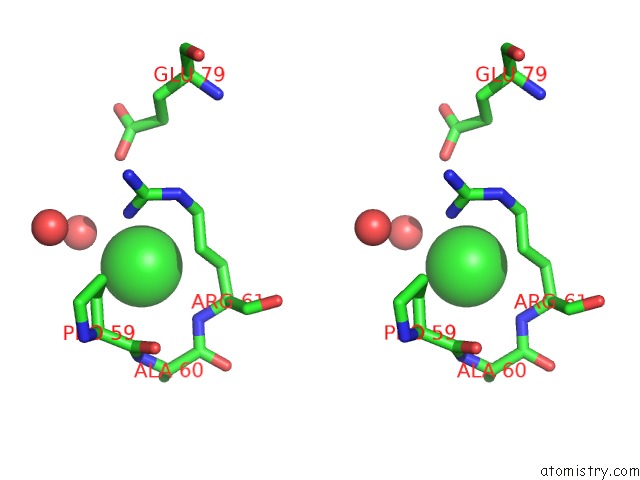
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Anti-Human Cytomegalovirus (Hcmv) Fab KE5 with Epitope Peptide Ad-2S1 within 5.0Å range:
|
Chlorine binding site 2 out of 5 in 4hha
Go back to
Chlorine binding site 2 out
of 5 in the Anti-Human Cytomegalovirus (Hcmv) Fab KE5 with Epitope Peptide Ad-2S1
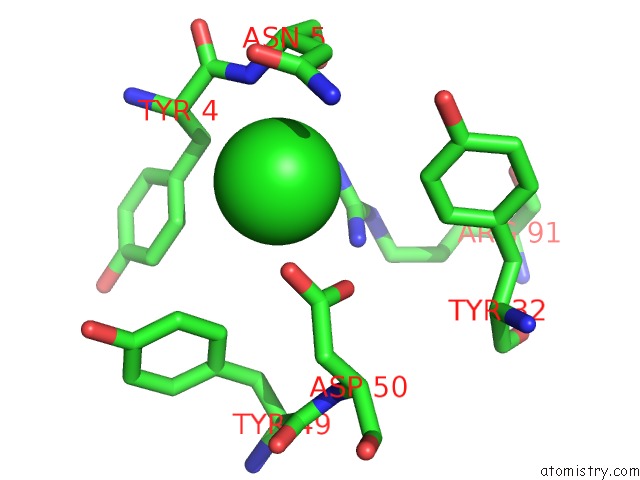
Mono view
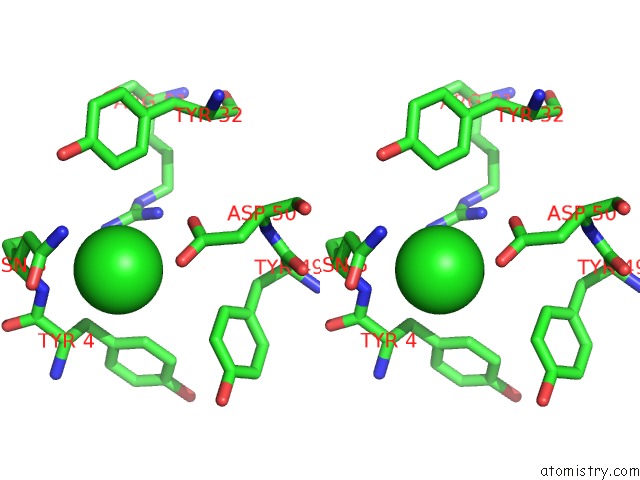
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Anti-Human Cytomegalovirus (Hcmv) Fab KE5 with Epitope Peptide Ad-2S1 within 5.0Å range:
|
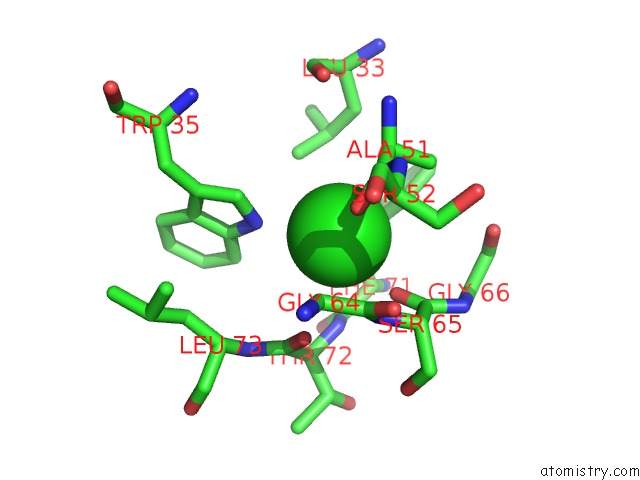
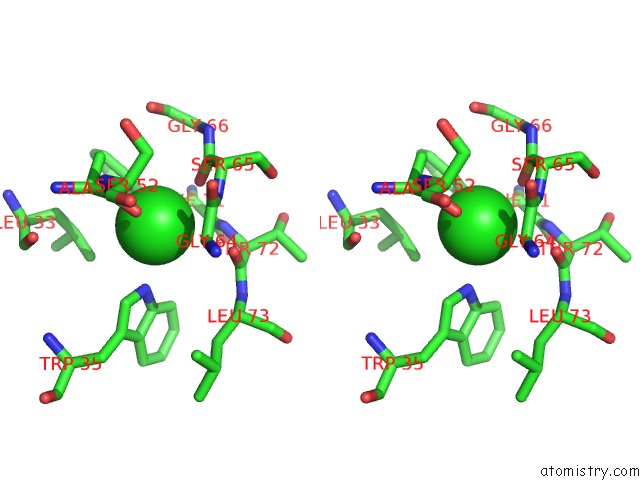
Chlorine binding site 3 out of 5 in 4hha
Go back to
Chlorine binding site 3 out
of 5 in the Anti-Human Cytomegalovirus (Hcmv) Fab KE5 with Epitope Peptide Ad-2S1
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Anti-Human Cytomegalovirus (Hcmv) Fab KE5 with Epitope Peptide Ad-2S1 within 5.0Å range:
|
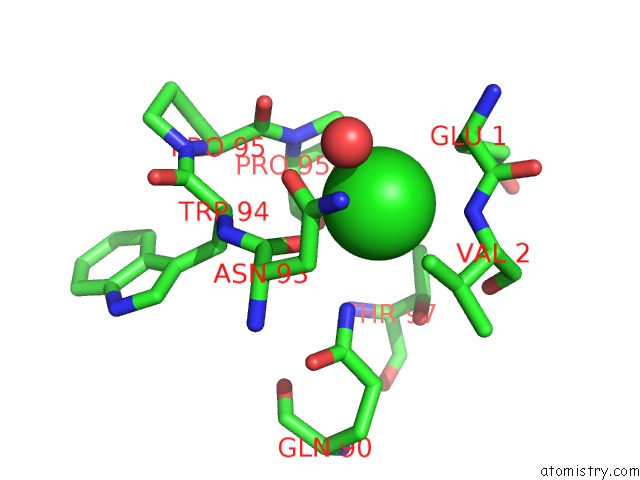
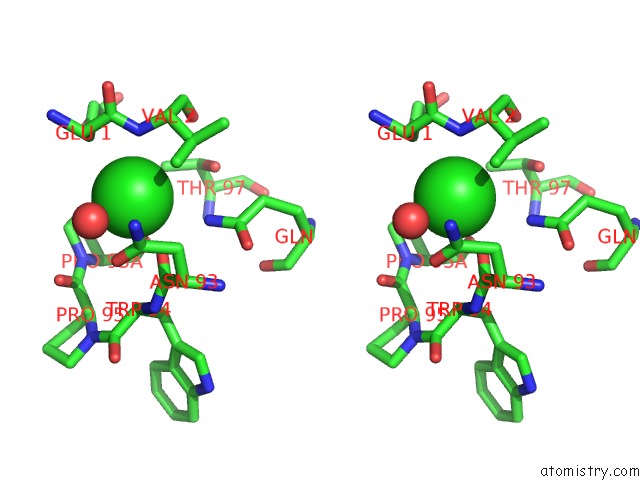
Chlorine binding site 4 out of 5 in 4hha
Go back to
Chlorine binding site 4 out
of 5 in the Anti-Human Cytomegalovirus (Hcmv) Fab KE5 with Epitope Peptide Ad-2S1
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Anti-Human Cytomegalovirus (Hcmv) Fab KE5 with Epitope Peptide Ad-2S1 within 5.0Å range:
|
Chlorine binding site 5 out of 5 in 4hha
Go back to
Chlorine binding site 5 out
of 5 in the Anti-Human Cytomegalovirus (Hcmv) Fab KE5 with Epitope Peptide Ad-2S1
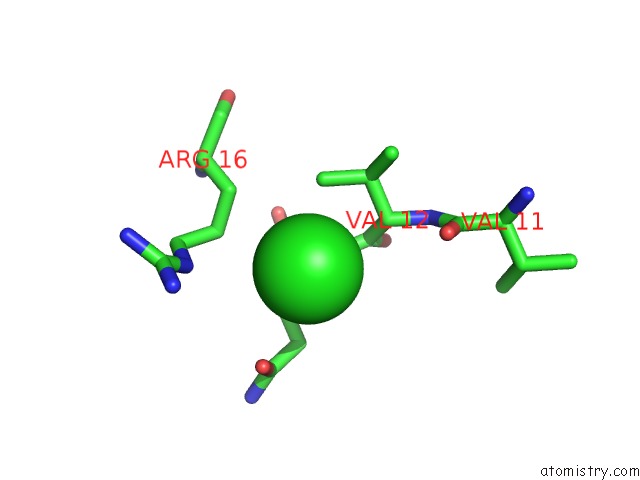
Mono view
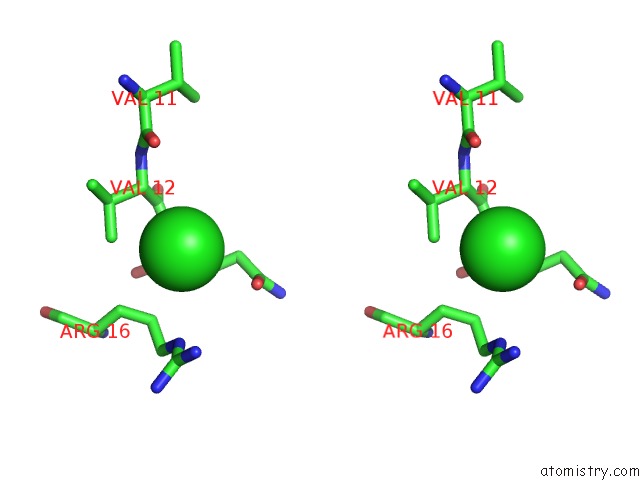
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Anti-Human Cytomegalovirus (Hcmv) Fab KE5 with Epitope Peptide Ad-2S1 within 5.0Å range:
|
Reference:
S.Bryson,
L.Risnes,
S.Damgupta,
C.A.Thomson,
J.W.Schrader,
E.F.Pai.
Multitasking Immunoglobulin V-Genes and Somatic Diversity in CDR3 Loops Generate Binding Sites For Chemically Distinct Antigens From Bacterial and Viral Pathogens. To Be Published.
Page generated: Sun Jul 21 15:50:25 2024
Last articles
Cl in 2WOCCl in 2WOD
Cl in 2WM2
Cl in 2WO4
Cl in 2WNY
Cl in 2WNQ
Cl in 2WMZ
Cl in 2WNT
Cl in 2WNS
Cl in 2WLJ