Chlorine »
PDB 4ibf-4iij »
4ieh »
Chlorine in PDB 4ieh: Crystal Structure of Human Bcl-2 in Complex with A Small Molecule Inhibitor Targeting Bcl-2 BH3 Domain Interactions
Protein crystallography data
The structure of Crystal Structure of Human Bcl-2 in Complex with A Small Molecule Inhibitor Targeting Bcl-2 BH3 Domain Interactions, PDB code: 4ieh
was solved by
X.Xie,
R.Kulathila,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 45.74 / 2.10 |
| Space group | P 43 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 73.529, 73.529, 96.205, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17.9 / 21.8 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of Human Bcl-2 in Complex with A Small Molecule Inhibitor Targeting Bcl-2 BH3 Domain Interactions
(pdb code 4ieh). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the Crystal Structure of Human Bcl-2 in Complex with A Small Molecule Inhibitor Targeting Bcl-2 BH3 Domain Interactions, PDB code: 4ieh:
In total only one binding site of Chlorine was determined in the Crystal Structure of Human Bcl-2 in Complex with A Small Molecule Inhibitor Targeting Bcl-2 BH3 Domain Interactions, PDB code: 4ieh:
Chlorine binding site 1 out of 1 in 4ieh
Go back to
Chlorine binding site 1 out
of 1 in the Crystal Structure of Human Bcl-2 in Complex with A Small Molecule Inhibitor Targeting Bcl-2 BH3 Domain Interactions
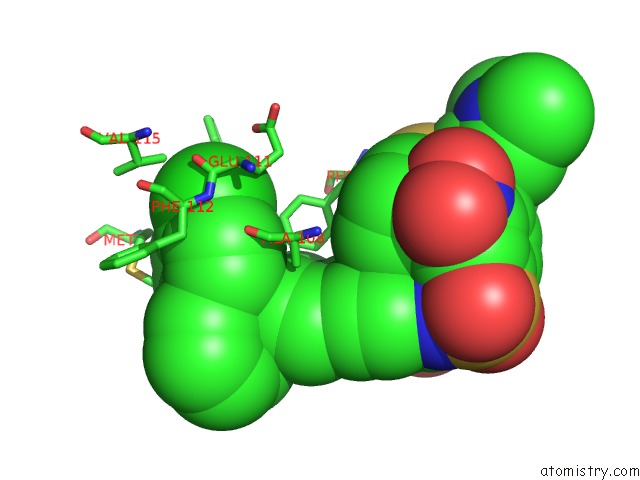
Mono view
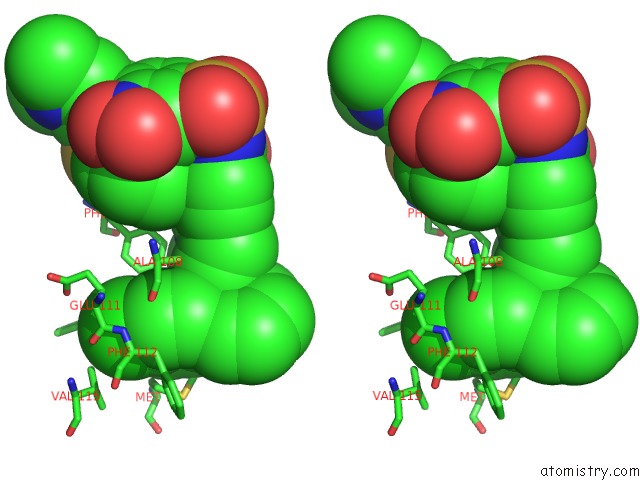
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of Human Bcl-2 in Complex with A Small Molecule Inhibitor Targeting Bcl-2 BH3 Domain Interactions within 5.0Å range:
|
Reference:
B.B.Toure,
K.Miller-Moslin,
N.Yusuff,
L.Perez,
M.Dore,
C.Joud,
W.Michael,
L.Dipietro,
S.Van Der Plas,
M.Mcewan,
F.Lenoir,
M.Hoe,
R.Karki,
C.Springer,
J.Sullivan,
K.Levine,
C.Fiorilla,
X.Xie,
R.Kulathila,
K.Herlihy,
D.Porter,
M.Visser.
The Role of the Acidity of N-Heteroaryl Sulfonamides As Inhibitors of Bcl-2 Family Protein-Protein Interactions. Acs Med Chem Lett V. 4 186 2013.
ISSN: ISSN 1948-5875
PubMed: 24900652
DOI: 10.1021/ML300321D
Page generated: Fri Jul 11 16:48:45 2025
ISSN: ISSN 1948-5875
PubMed: 24900652
DOI: 10.1021/ML300321D
Last articles
Cl in 4XFZCl in 4XFY
Cl in 4XF5
Cl in 4XFW
Cl in 4XFX
Cl in 4XFG
Cl in 4XFF
Cl in 4XEN
Cl in 4XFC
Cl in 4XFB