Chlorine »
PDB 4io4-4ixu »
4isu »
Chlorine in PDB 4isu: Crystal Structure of the GLUA2 Ligand-Binding Domain (S1S2J) in Complex with the Antagonist (2R)-Ikm-159 at 2.3A Resolution.
Protein crystallography data
The structure of Crystal Structure of the GLUA2 Ligand-Binding Domain (S1S2J) in Complex with the Antagonist (2R)-Ikm-159 at 2.3A Resolution., PDB code: 4isu
was solved by
L.Juknaite,
K.Frydenvang,
J.S.Kastrup,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 29.15 / 2.30 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 62.514, 88.773, 194.456, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 18.5 / 25.8 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of the GLUA2 Ligand-Binding Domain (S1S2J) in Complex with the Antagonist (2R)-Ikm-159 at 2.3A Resolution.
(pdb code 4isu). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 8 binding sites of Chlorine where determined in the Crystal Structure of the GLUA2 Ligand-Binding Domain (S1S2J) in Complex with the Antagonist (2R)-Ikm-159 at 2.3A Resolution., PDB code: 4isu:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
In total 8 binding sites of Chlorine where determined in the Crystal Structure of the GLUA2 Ligand-Binding Domain (S1S2J) in Complex with the Antagonist (2R)-Ikm-159 at 2.3A Resolution., PDB code: 4isu:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
Chlorine binding site 1 out of 8 in 4isu
Go back to
Chlorine binding site 1 out
of 8 in the Crystal Structure of the GLUA2 Ligand-Binding Domain (S1S2J) in Complex with the Antagonist (2R)-Ikm-159 at 2.3A Resolution.
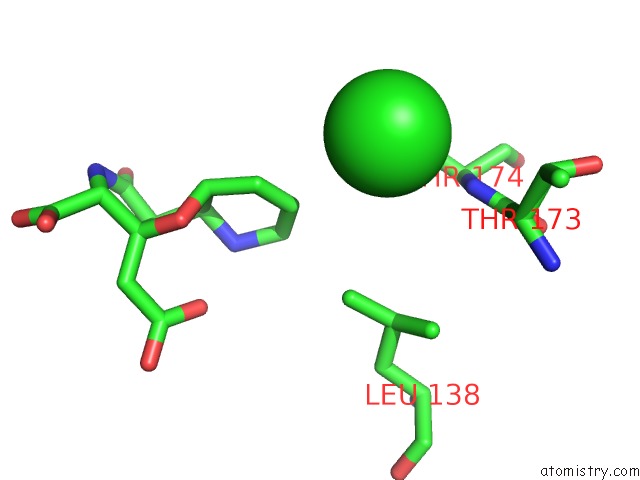
Mono view
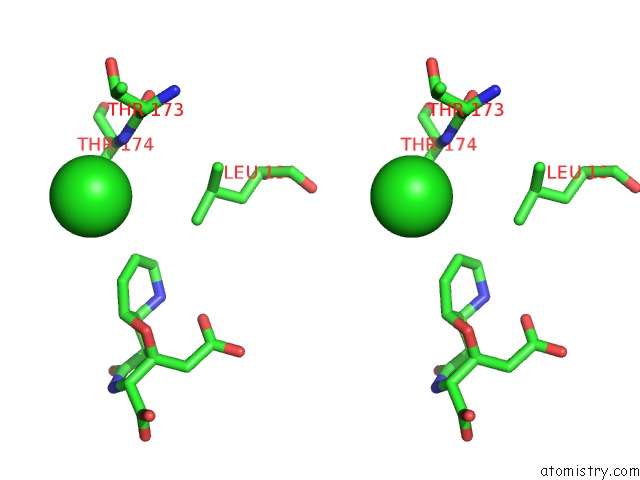
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of the GLUA2 Ligand-Binding Domain (S1S2J) in Complex with the Antagonist (2R)-Ikm-159 at 2.3A Resolution. within 5.0Å range:
|
Chlorine binding site 2 out of 8 in 4isu
Go back to
Chlorine binding site 2 out
of 8 in the Crystal Structure of the GLUA2 Ligand-Binding Domain (S1S2J) in Complex with the Antagonist (2R)-Ikm-159 at 2.3A Resolution.
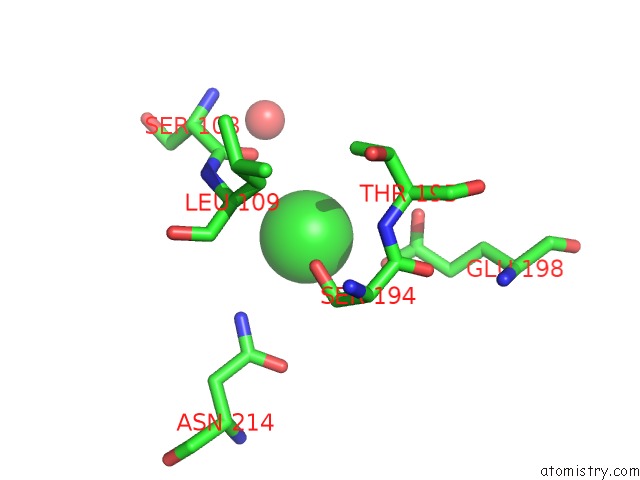
Mono view
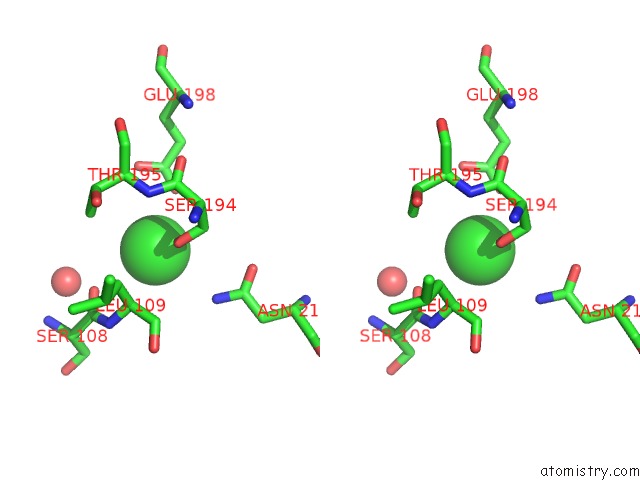
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of the GLUA2 Ligand-Binding Domain (S1S2J) in Complex with the Antagonist (2R)-Ikm-159 at 2.3A Resolution. within 5.0Å range:
|
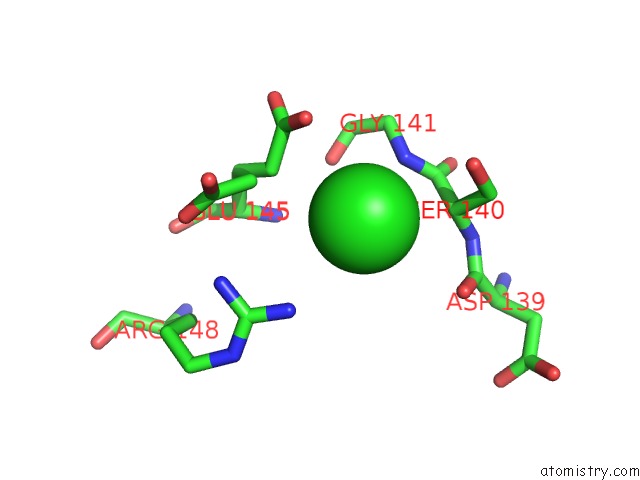
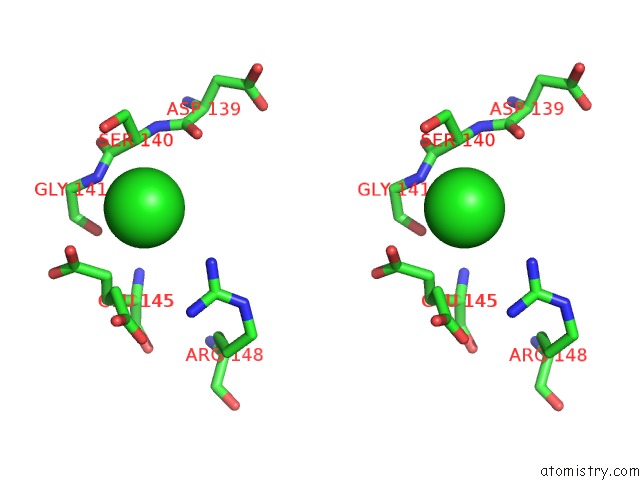
Chlorine binding site 3 out of 8 in 4isu
Go back to
Chlorine binding site 3 out
of 8 in the Crystal Structure of the GLUA2 Ligand-Binding Domain (S1S2J) in Complex with the Antagonist (2R)-Ikm-159 at 2.3A Resolution.
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Crystal Structure of the GLUA2 Ligand-Binding Domain (S1S2J) in Complex with the Antagonist (2R)-Ikm-159 at 2.3A Resolution. within 5.0Å range:
|
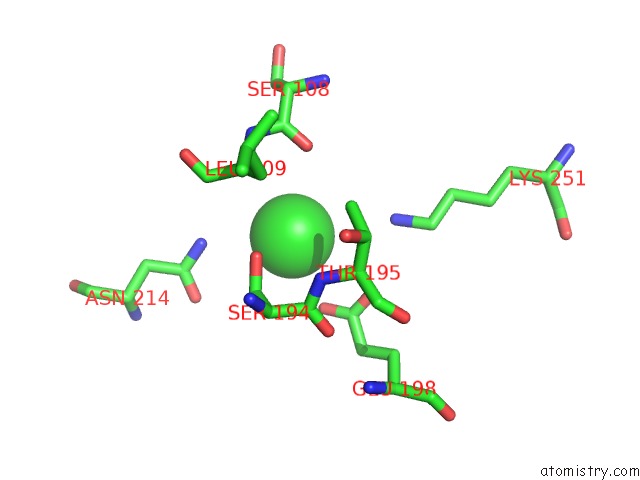
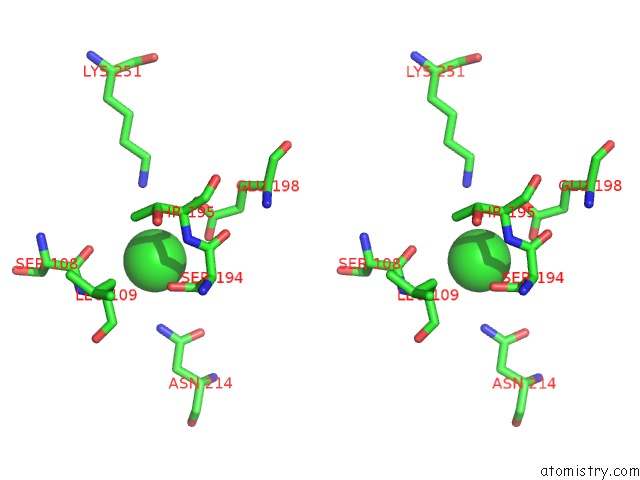
Chlorine binding site 4 out of 8 in 4isu
Go back to
Chlorine binding site 4 out
of 8 in the Crystal Structure of the GLUA2 Ligand-Binding Domain (S1S2J) in Complex with the Antagonist (2R)-Ikm-159 at 2.3A Resolution.
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Crystal Structure of the GLUA2 Ligand-Binding Domain (S1S2J) in Complex with the Antagonist (2R)-Ikm-159 at 2.3A Resolution. within 5.0Å range:
|
Chlorine binding site 5 out of 8 in 4isu
Go back to
Chlorine binding site 5 out
of 8 in the Crystal Structure of the GLUA2 Ligand-Binding Domain (S1S2J) in Complex with the Antagonist (2R)-Ikm-159 at 2.3A Resolution.
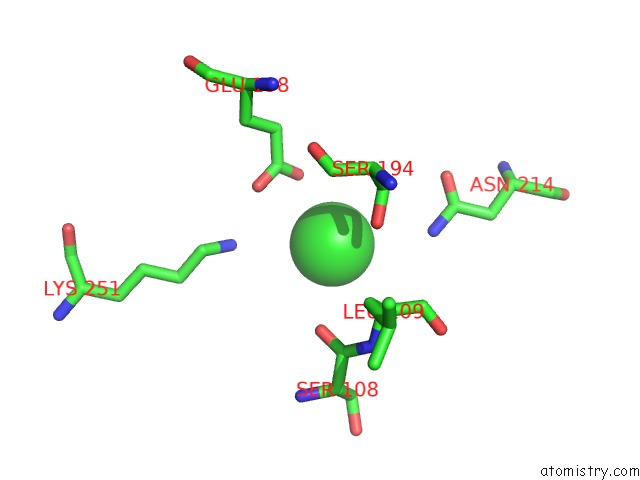
Mono view
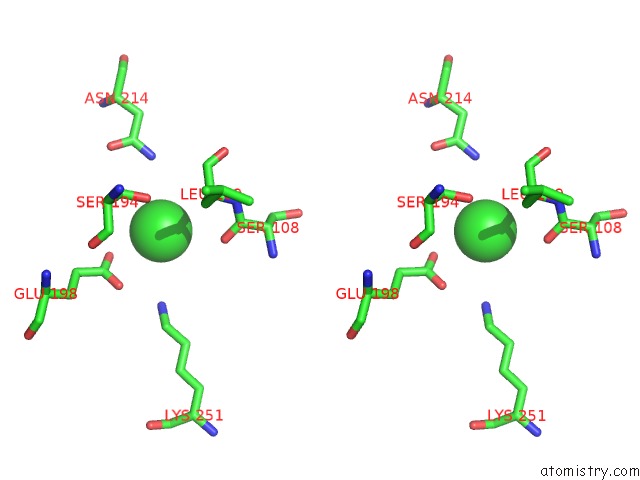
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Crystal Structure of the GLUA2 Ligand-Binding Domain (S1S2J) in Complex with the Antagonist (2R)-Ikm-159 at 2.3A Resolution. within 5.0Å range:
|
Chlorine binding site 6 out of 8 in 4isu
Go back to
Chlorine binding site 6 out
of 8 in the Crystal Structure of the GLUA2 Ligand-Binding Domain (S1S2J) in Complex with the Antagonist (2R)-Ikm-159 at 2.3A Resolution.
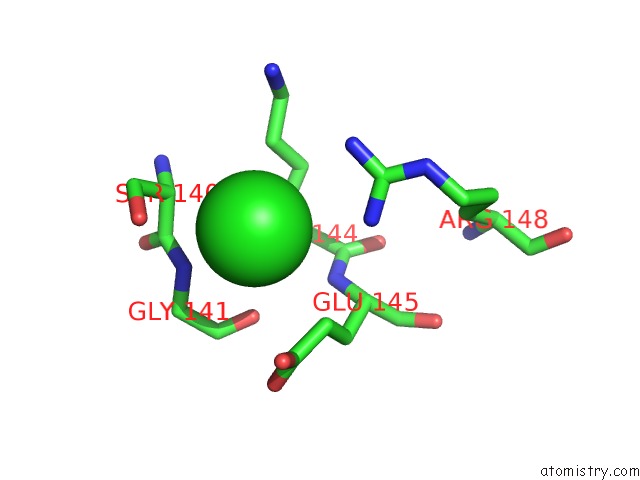
Mono view
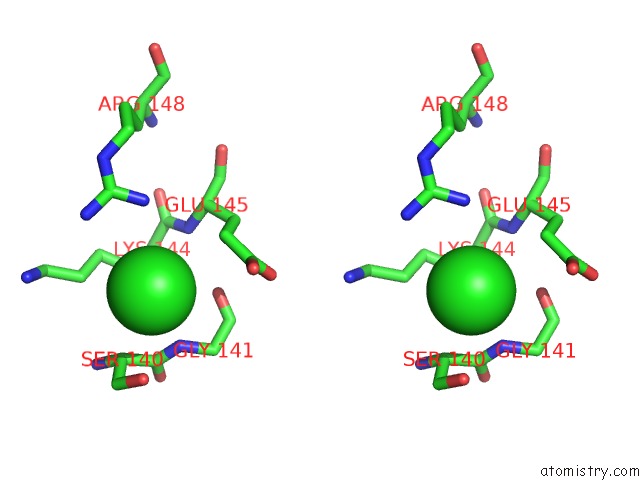
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 6 of Crystal Structure of the GLUA2 Ligand-Binding Domain (S1S2J) in Complex with the Antagonist (2R)-Ikm-159 at 2.3A Resolution. within 5.0Å range:
|
Chlorine binding site 7 out of 8 in 4isu
Go back to
Chlorine binding site 7 out
of 8 in the Crystal Structure of the GLUA2 Ligand-Binding Domain (S1S2J) in Complex with the Antagonist (2R)-Ikm-159 at 2.3A Resolution.
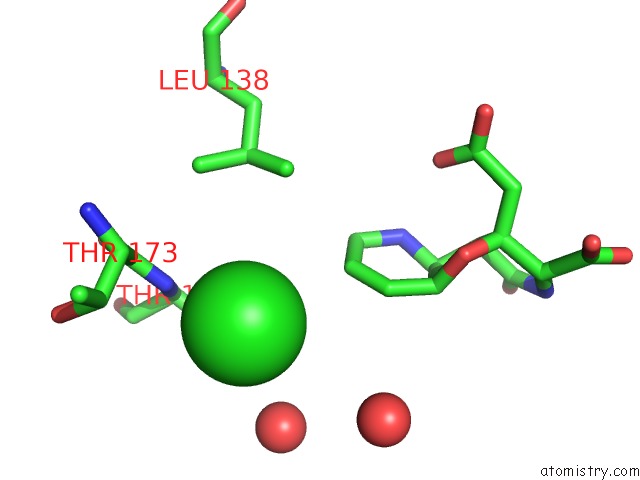
Mono view
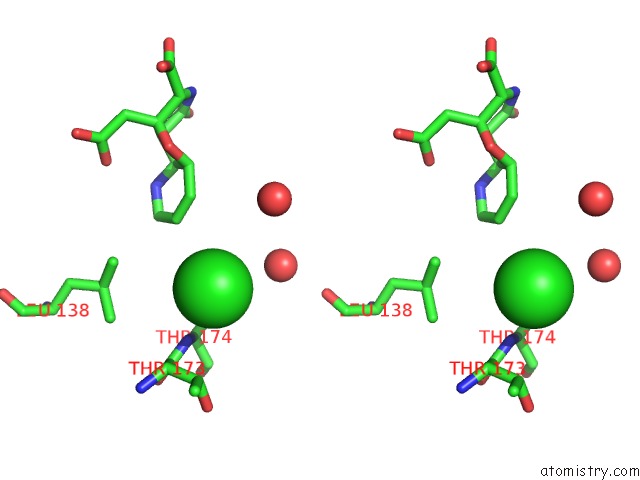
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 7 of Crystal Structure of the GLUA2 Ligand-Binding Domain (S1S2J) in Complex with the Antagonist (2R)-Ikm-159 at 2.3A Resolution. within 5.0Å range:
|
Chlorine binding site 8 out of 8 in 4isu
Go back to
Chlorine binding site 8 out
of 8 in the Crystal Structure of the GLUA2 Ligand-Binding Domain (S1S2J) in Complex with the Antagonist (2R)-Ikm-159 at 2.3A Resolution.
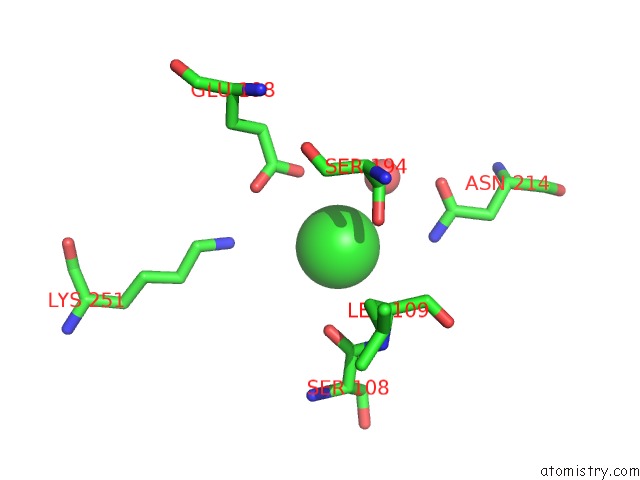
Mono view
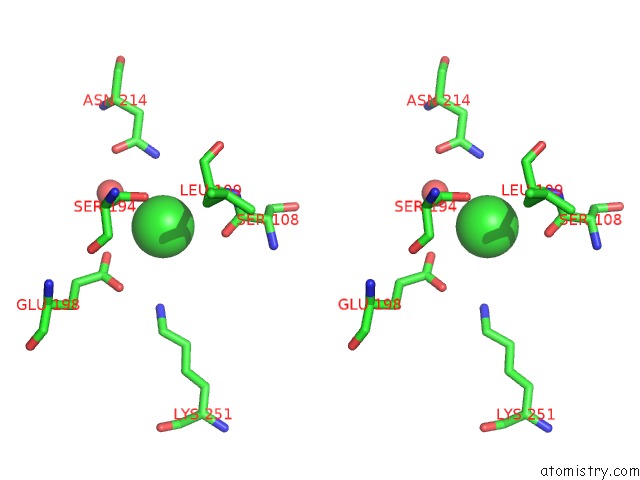
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 8 of Crystal Structure of the GLUA2 Ligand-Binding Domain (S1S2J) in Complex with the Antagonist (2R)-Ikm-159 at 2.3A Resolution. within 5.0Å range:
|
Reference:
L.Juknaite,
Y.Sugamata,
K.Tokiwa,
Y.Ishikawa,
S.Takamizawa,
A.Eng,
R.Sakai,
D.S.Pickering,
K.Frydenvang,
G.T.Swanson,
J.S.Kastrup,
M.Oikawa.
Studies on An (S)-2-Amino-3-(3-Hydroxy-5-Methyl-4-Isoxazolyl)Propionic Acid (Ampa) Receptor Antagonist Ikm-159: Asymmetric Synthesis, Neuroactivity, and Structural Characterization. J.Med.Chem. V. 56 2283 2013.
ISSN: ISSN 0022-2623
PubMed: 23432124
DOI: 10.1021/JM301590Z
Page generated: Fri Jul 11 17:04:25 2025
ISSN: ISSN 0022-2623
PubMed: 23432124
DOI: 10.1021/JM301590Z
Last articles
F in 7MRCF in 7MPF
F in 7MR7
F in 7MPB
F in 7MR6
F in 7MNG
F in 7MR5
F in 7MON
F in 7MOG
F in 7MOO