Chlorine »
PDB 4kbn-4kmz »
4klm »
Chlorine in PDB 4klm: Dna Polymerase Beta Matched Product Complex with MG2+, 11 H
Enzymatic activity of Dna Polymerase Beta Matched Product Complex with MG2+, 11 H
All present enzymatic activity of Dna Polymerase Beta Matched Product Complex with MG2+, 11 H:
2.7.7.7;
2.7.7.7;
Protein crystallography data
The structure of Dna Polymerase Beta Matched Product Complex with MG2+, 11 H, PDB code: 4klm
was solved by
B.D.Freudenthal,
W.A.Beard,
D.D.Shock,
S.H.Wilson,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 23.83 / 1.75 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 50.588, 79.760, 55.338, 90.00, 107.70, 90.00 |
| R / Rfree (%) | 18.8 / 23.9 |
Other elements in 4klm:
The structure of Dna Polymerase Beta Matched Product Complex with MG2+, 11 H also contains other interesting chemical elements:
| Magnesium | (Mg) | 1 atom |
| Sodium | (Na) | 3 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Dna Polymerase Beta Matched Product Complex with MG2+, 11 H
(pdb code 4klm). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Dna Polymerase Beta Matched Product Complex with MG2+, 11 H, PDB code: 4klm:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Dna Polymerase Beta Matched Product Complex with MG2+, 11 H, PDB code: 4klm:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 4klm
Go back to
Chlorine binding site 1 out
of 2 in the Dna Polymerase Beta Matched Product Complex with MG2+, 11 H
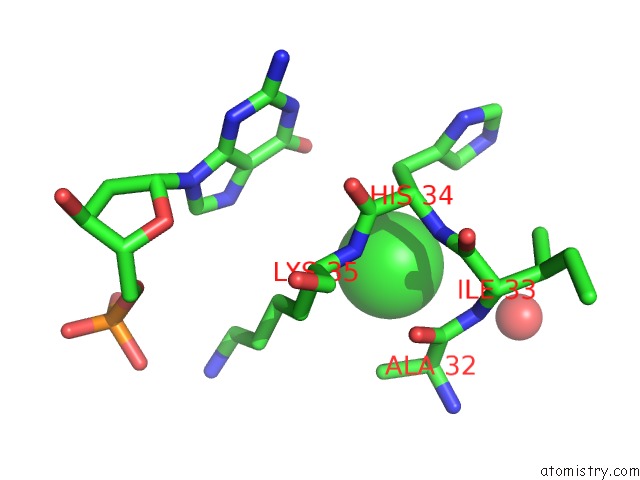
Mono view
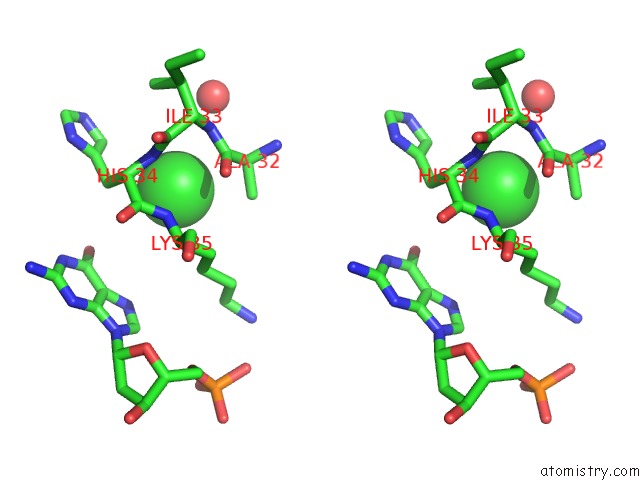
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Dna Polymerase Beta Matched Product Complex with MG2+, 11 H within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 4klm
Go back to
Chlorine binding site 2 out
of 2 in the Dna Polymerase Beta Matched Product Complex with MG2+, 11 H
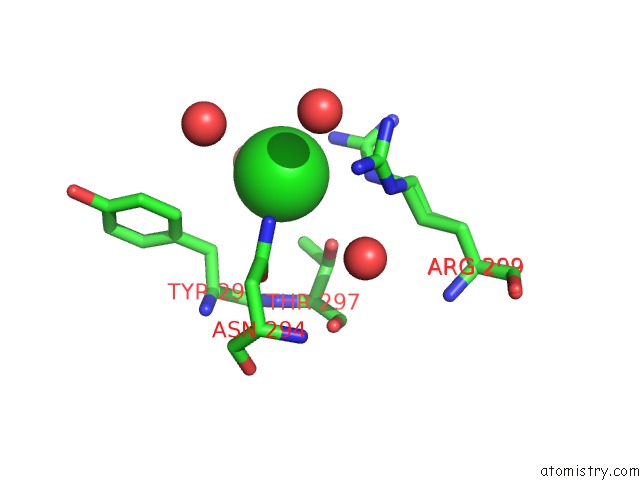
Mono view
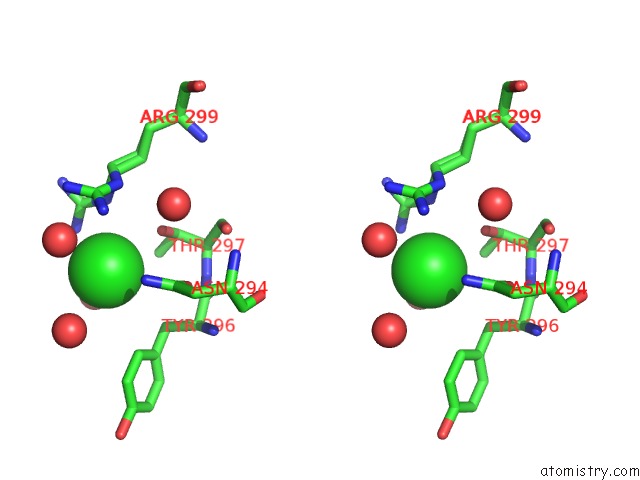
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Dna Polymerase Beta Matched Product Complex with MG2+, 11 H within 5.0Å range:
|
Reference:
B.D.Freudenthal,
W.A.Beard,
D.D.Shock,
S.H.Wilson.
Observing A Dna Polymerase Choose Right From Wrong. Cell(Cambridge,Mass.) V. 154 157 2013.
ISSN: ISSN 0092-8674
PubMed: 23827680
DOI: 10.1016/J.CELL.2013.05.048
Page generated: Fri Jul 11 17:48:34 2025
ISSN: ISSN 0092-8674
PubMed: 23827680
DOI: 10.1016/J.CELL.2013.05.048
Last articles
F in 7QRAF in 7QRB
F in 7QRC
F in 7QR9
F in 7QQ6
F in 7QPZ
F in 7QPY
F in 7QN0
F in 7QN4
F in 7QN1