Chlorine »
PDB 4kbn-4kmz »
4kmw »
Chlorine in PDB 4kmw: Structure of the Y34N Mutant of Dehaloperoxidase-Hemoglobin A From Amphitrite Ornata with 2,4,6-Trichlorophenol
Protein crystallography data
The structure of Structure of the Y34N Mutant of Dehaloperoxidase-Hemoglobin A From Amphitrite Ornata with 2,4,6-Trichlorophenol, PDB code: 4kmw
was solved by
C.Wang,
L.Lovelace,
L.Lebioda,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 47.78 / 1.79 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 58.553, 67.259, 67.890, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 14.6 / 23.2 |
Other elements in 4kmw:
The structure of Structure of the Y34N Mutant of Dehaloperoxidase-Hemoglobin A From Amphitrite Ornata with 2,4,6-Trichlorophenol also contains other interesting chemical elements:
| Iron | (Fe) | 2 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Structure of the Y34N Mutant of Dehaloperoxidase-Hemoglobin A From Amphitrite Ornata with 2,4,6-Trichlorophenol
(pdb code 4kmw). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 6 binding sites of Chlorine where determined in the Structure of the Y34N Mutant of Dehaloperoxidase-Hemoglobin A From Amphitrite Ornata with 2,4,6-Trichlorophenol, PDB code: 4kmw:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Chlorine where determined in the Structure of the Y34N Mutant of Dehaloperoxidase-Hemoglobin A From Amphitrite Ornata with 2,4,6-Trichlorophenol, PDB code: 4kmw:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6;
Chlorine binding site 1 out of 6 in 4kmw
Go back to
Chlorine binding site 1 out
of 6 in the Structure of the Y34N Mutant of Dehaloperoxidase-Hemoglobin A From Amphitrite Ornata with 2,4,6-Trichlorophenol
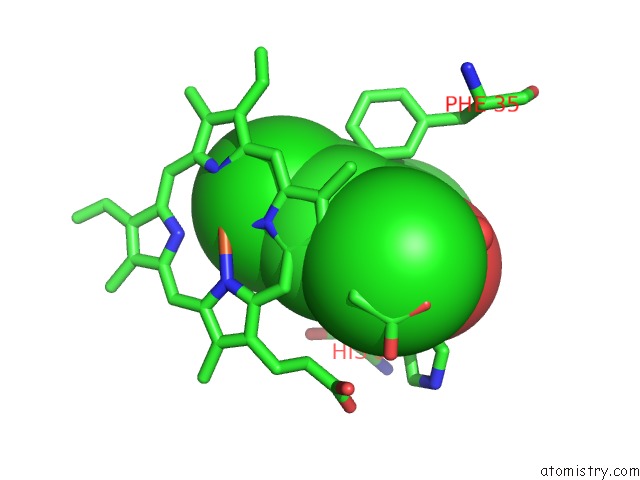
Mono view
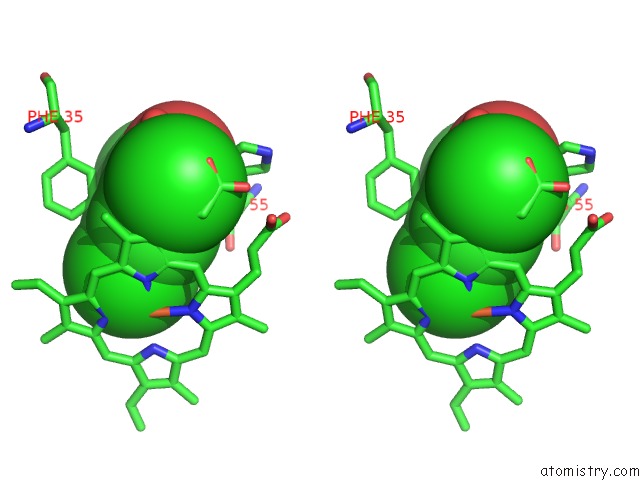
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Structure of the Y34N Mutant of Dehaloperoxidase-Hemoglobin A From Amphitrite Ornata with 2,4,6-Trichlorophenol within 5.0Å range:
|
Chlorine binding site 2 out of 6 in 4kmw
Go back to
Chlorine binding site 2 out
of 6 in the Structure of the Y34N Mutant of Dehaloperoxidase-Hemoglobin A From Amphitrite Ornata with 2,4,6-Trichlorophenol
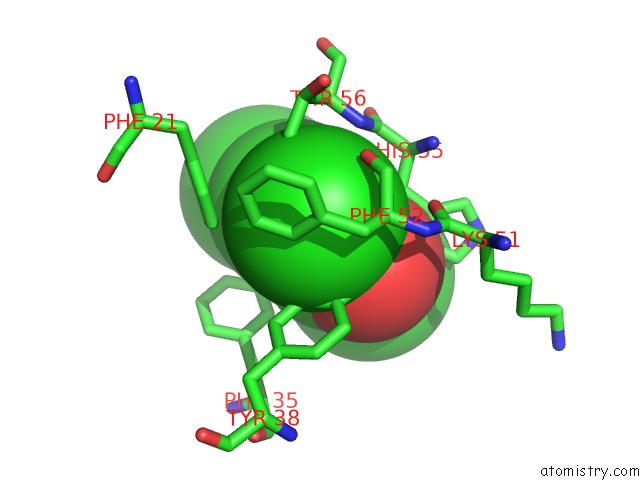
Mono view
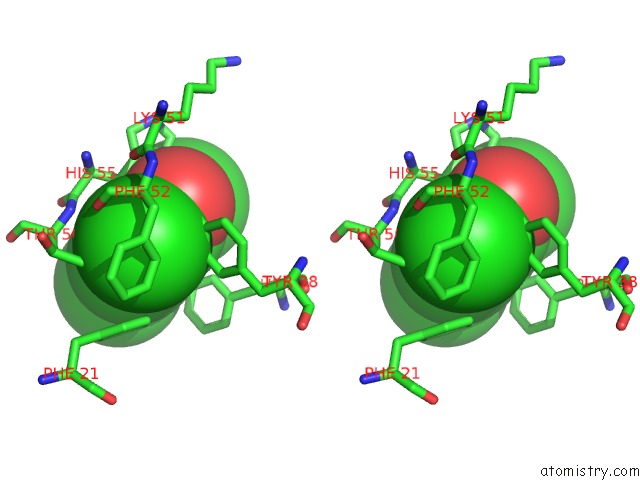
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Structure of the Y34N Mutant of Dehaloperoxidase-Hemoglobin A From Amphitrite Ornata with 2,4,6-Trichlorophenol within 5.0Å range:
|
Chlorine binding site 3 out of 6 in 4kmw
Go back to
Chlorine binding site 3 out
of 6 in the Structure of the Y34N Mutant of Dehaloperoxidase-Hemoglobin A From Amphitrite Ornata with 2,4,6-Trichlorophenol
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Structure of the Y34N Mutant of Dehaloperoxidase-Hemoglobin A From Amphitrite Ornata with 2,4,6-Trichlorophenol within 5.0Å range:
|
Chlorine binding site 4 out of 6 in 4kmw
Go back to
Chlorine binding site 4 out
of 6 in the Structure of the Y34N Mutant of Dehaloperoxidase-Hemoglobin A From Amphitrite Ornata with 2,4,6-Trichlorophenol
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Structure of the Y34N Mutant of Dehaloperoxidase-Hemoglobin A From Amphitrite Ornata with 2,4,6-Trichlorophenol within 5.0Å range:
|
Chlorine binding site 5 out of 6 in 4kmw
Go back to
Chlorine binding site 5 out
of 6 in the Structure of the Y34N Mutant of Dehaloperoxidase-Hemoglobin A From Amphitrite Ornata with 2,4,6-Trichlorophenol
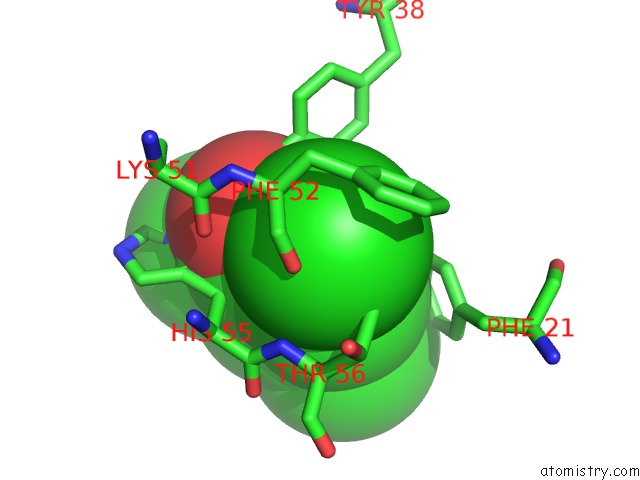
Mono view
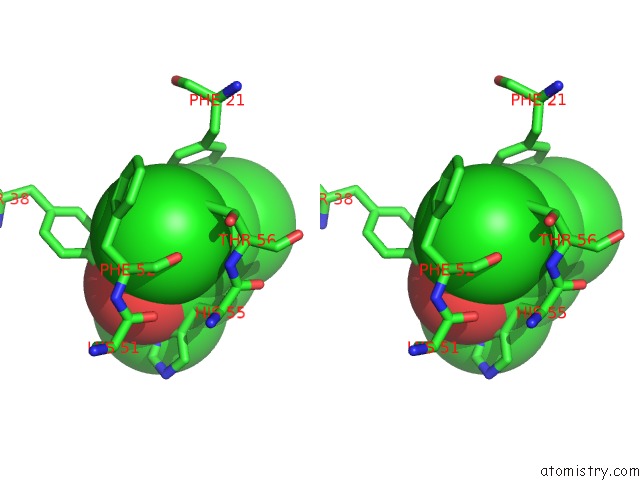
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Structure of the Y34N Mutant of Dehaloperoxidase-Hemoglobin A From Amphitrite Ornata with 2,4,6-Trichlorophenol within 5.0Å range:
|
Chlorine binding site 6 out of 6 in 4kmw
Go back to
Chlorine binding site 6 out
of 6 in the Structure of the Y34N Mutant of Dehaloperoxidase-Hemoglobin A From Amphitrite Ornata with 2,4,6-Trichlorophenol
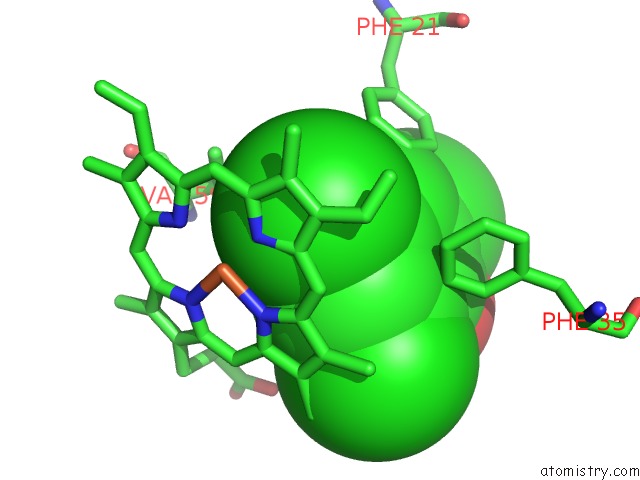
Mono view
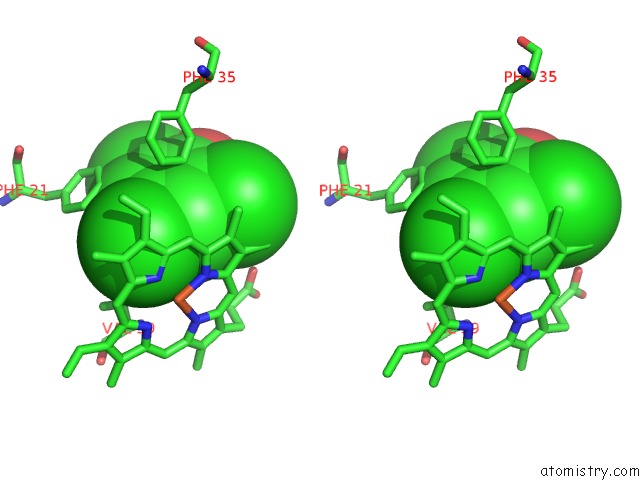
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 6 of Structure of the Y34N Mutant of Dehaloperoxidase-Hemoglobin A From Amphitrite Ornata with 2,4,6-Trichlorophenol within 5.0Å range:
|
Reference:
C.Wang,
L.L.Lovelace,
S.Sun,
J.H.Dawson,
L.Lebioda.
Complexes of Dual-Function Hemoglobin/Dehaloperoxidase with Substrate 2,4,6-Trichlorophenol Are Inhibitory and Indicate Binding of Halophenol to Compound I. Biochemistry V. 52 6203 2013.
ISSN: ISSN 0006-2960
PubMed: 23952341
DOI: 10.1021/BI400627W
Page generated: Fri Jul 11 17:49:23 2025
ISSN: ISSN 0006-2960
PubMed: 23952341
DOI: 10.1021/BI400627W
Last articles
Fe in 2J19Fe in 2J18
Fe in 2J0P
Fe in 2IW4
Fe in 2J0D
Fe in 2IV2
Fe in 2IVP
Fe in 2IVJ
Fe in 2ISA
Fe in 2IVI