Chlorine »
PDB 4m5h-4mdq »
4mdq »
Chlorine in PDB 4mdq: Structure of A Novel Submicromolar MDM2 Inhibitor
Protein crystallography data
The structure of Structure of A Novel Submicromolar MDM2 Inhibitor, PDB code: 4mdq
was solved by
M.Bista,
G.Popowicz,
T.A.Holak,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 46.35 / 2.12 |
| Space group | P 65 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 53.520, 53.520, 122.270, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 18.5 / 22.1 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Structure of A Novel Submicromolar MDM2 Inhibitor
(pdb code 4mdq). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Structure of A Novel Submicromolar MDM2 Inhibitor, PDB code: 4mdq:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Structure of A Novel Submicromolar MDM2 Inhibitor, PDB code: 4mdq:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 4mdq
Go back to
Chlorine binding site 1 out
of 2 in the Structure of A Novel Submicromolar MDM2 Inhibitor
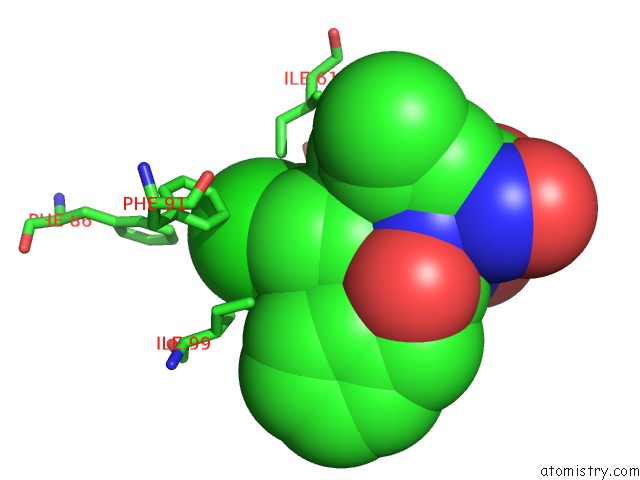
Mono view
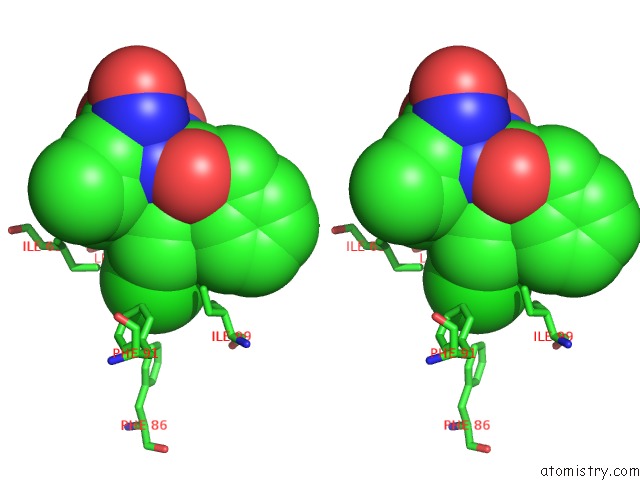
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Structure of A Novel Submicromolar MDM2 Inhibitor within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 4mdq
Go back to
Chlorine binding site 2 out
of 2 in the Structure of A Novel Submicromolar MDM2 Inhibitor
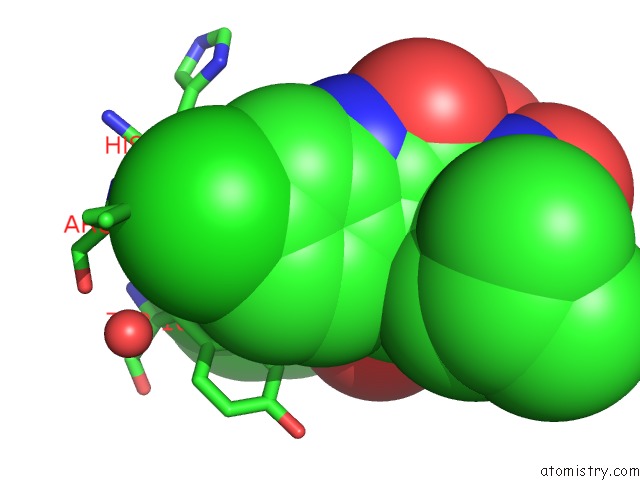
Mono view
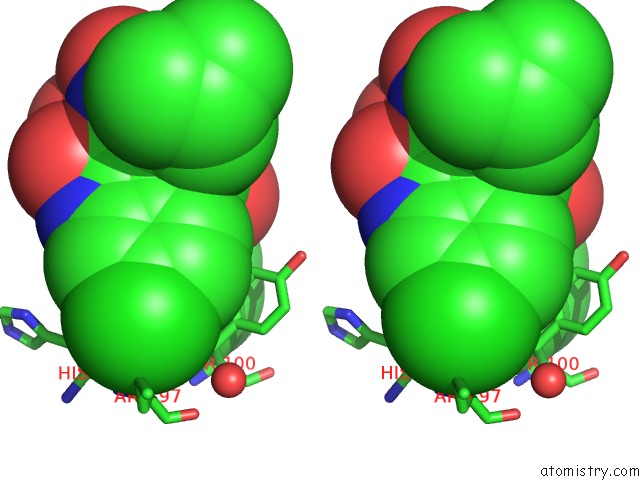
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Structure of A Novel Submicromolar MDM2 Inhibitor within 5.0Å range:
|
Reference:
M.Bista,
S.Wolf,
K.Khoury,
K.Kowalska,
Y.Huang,
E.Wrona,
M.Arciniega,
G.M.Popowicz,
T.A.Holak,
A.Domling.
Transient Protein States in Designing Inhibitors of the MDM2-P53 Interaction. Structure V. 21 2143 2013.
ISSN: ISSN 0969-2126
PubMed: 24207125
DOI: 10.1016/J.STR.2013.09.006
Page generated: Fri Jul 11 19:02:17 2025
ISSN: ISSN 0969-2126
PubMed: 24207125
DOI: 10.1016/J.STR.2013.09.006
Last articles
F in 7QG3F in 7QHN
F in 7Q6P
F in 7QBZ
F in 7QC0
F in 7QAV
F in 7QA0
F in 7QA3
F in 7Q85
F in 7Q7R