Chlorine »
PDB 4mlb-4ms4 »
4mlo »
Chlorine in PDB 4mlo: 1.65A Resolution Structure of Toxt From Vibrio Cholerae (P21 Form)
Protein crystallography data
The structure of 1.65A Resolution Structure of Toxt From Vibrio Cholerae (P21 Form), PDB code: 4mlo
was solved by
S.Lovell,
G.Wehmeyer,
K.P.Battaile,
J.Li,
S.Egan,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 39.74 / 1.65 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 47.337, 39.410, 80.238, 90.00, 97.94, 90.00 |
| R / Rfree (%) | 16.8 / 19.4 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the 1.65A Resolution Structure of Toxt From Vibrio Cholerae (P21 Form)
(pdb code 4mlo). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 3 binding sites of Chlorine where determined in the 1.65A Resolution Structure of Toxt From Vibrio Cholerae (P21 Form), PDB code: 4mlo:
Jump to Chlorine binding site number: 1; 2; 3;
In total 3 binding sites of Chlorine where determined in the 1.65A Resolution Structure of Toxt From Vibrio Cholerae (P21 Form), PDB code: 4mlo:
Jump to Chlorine binding site number: 1; 2; 3;
Chlorine binding site 1 out of 3 in 4mlo
Go back to
Chlorine binding site 1 out
of 3 in the 1.65A Resolution Structure of Toxt From Vibrio Cholerae (P21 Form)
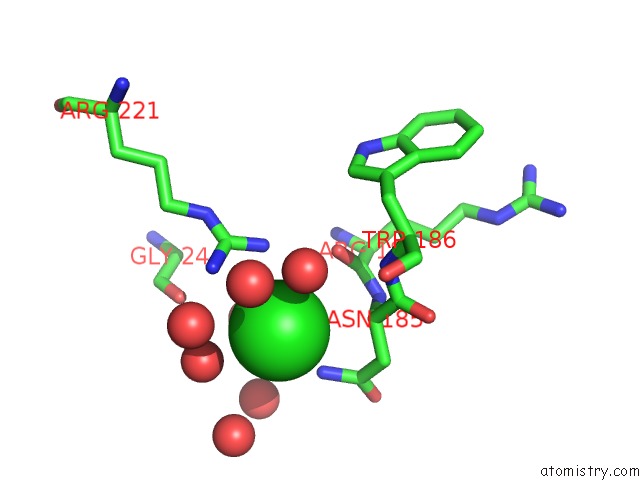
Mono view
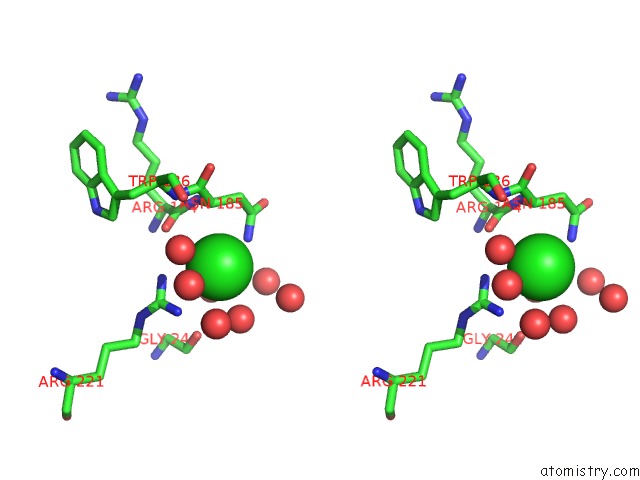
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of 1.65A Resolution Structure of Toxt From Vibrio Cholerae (P21 Form) within 5.0Å range:
|
Chlorine binding site 2 out of 3 in 4mlo
Go back to
Chlorine binding site 2 out
of 3 in the 1.65A Resolution Structure of Toxt From Vibrio Cholerae (P21 Form)
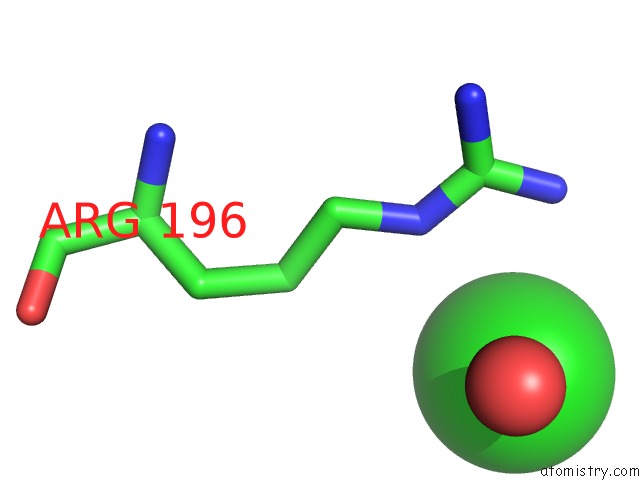
Mono view
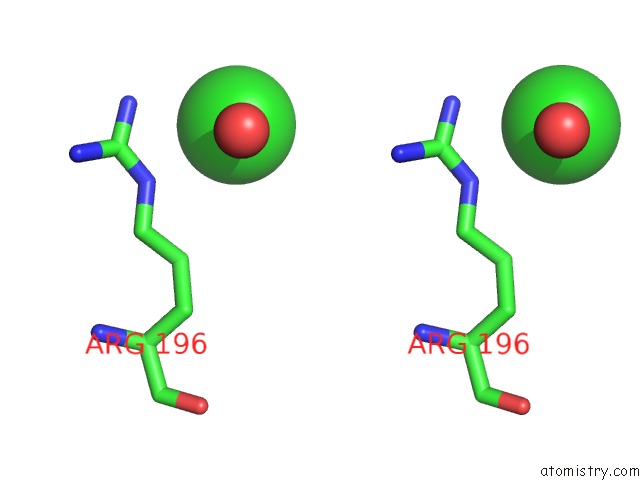
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of 1.65A Resolution Structure of Toxt From Vibrio Cholerae (P21 Form) within 5.0Å range:
|
Chlorine binding site 3 out of 3 in 4mlo
Go back to
Chlorine binding site 3 out
of 3 in the 1.65A Resolution Structure of Toxt From Vibrio Cholerae (P21 Form)
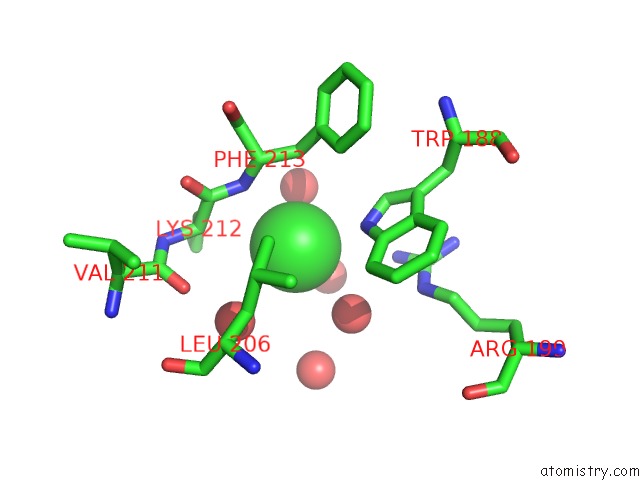
Mono view
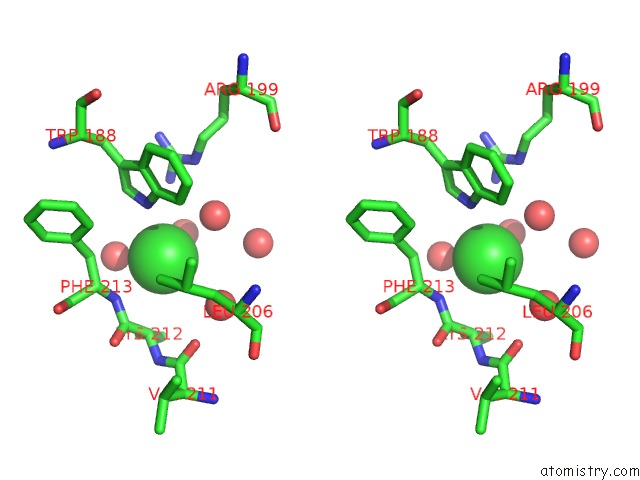
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of 1.65A Resolution Structure of Toxt From Vibrio Cholerae (P21 Form) within 5.0Å range:
|
Reference:
J.Li,
G.Wehmeyer,
S.Lovell,
K.P.Battaile,
S.M.Egan.
1.65 Angstrom Resolution Structure of the Arac-Family Transcriptional Activator Toxt From Vibrio Cholerae. Acta Crystallogr F Struct V. 72 726 2016BIOL Commun.
ISSN: ESSN 1744-3091
PubMed: 27599865
DOI: 10.1107/S2053230X1601298X
Page generated: Fri Jul 11 19:12:40 2025
ISSN: ESSN 1744-3091
PubMed: 27599865
DOI: 10.1107/S2053230X1601298X
Last articles
Cl in 5DVHCl in 5DVR
Cl in 5DU5
Cl in 5DVA
Cl in 5DU9
Cl in 5DUA
Cl in 5DTT
Cl in 5DTQ
Cl in 5DTM
Cl in 5DTR