Chlorine »
PDB 4myx-4n99 »
4n6x »
Chlorine in PDB 4n6x: Crystal Structure of the Chemokine Receptor CXCR2 in Complex with the First Pdz Domain of NHERF1
Protein crystallography data
The structure of Crystal Structure of the Chemokine Receptor CXCR2 in Complex with the First Pdz Domain of NHERF1, PDB code: 4n6x
was solved by
G.Lu,
Y.Wu,
Y.Jiang,
J.Brunzelle,
N.Sirinupong,
C.Li,
Z.Yang,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 26.30 / 1.05 |
| Space group | P 31 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 50.380, 50.380, 65.940, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 14.8 / 16.3 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of the Chemokine Receptor CXCR2 in Complex with the First Pdz Domain of NHERF1
(pdb code 4n6x). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 4 binding sites of Chlorine where determined in the Crystal Structure of the Chemokine Receptor CXCR2 in Complex with the First Pdz Domain of NHERF1, PDB code: 4n6x:
Jump to Chlorine binding site number: 1; 2; 3; 4;
In total 4 binding sites of Chlorine where determined in the Crystal Structure of the Chemokine Receptor CXCR2 in Complex with the First Pdz Domain of NHERF1, PDB code: 4n6x:
Jump to Chlorine binding site number: 1; 2; 3; 4;
Chlorine binding site 1 out of 4 in 4n6x
Go back to
Chlorine binding site 1 out
of 4 in the Crystal Structure of the Chemokine Receptor CXCR2 in Complex with the First Pdz Domain of NHERF1
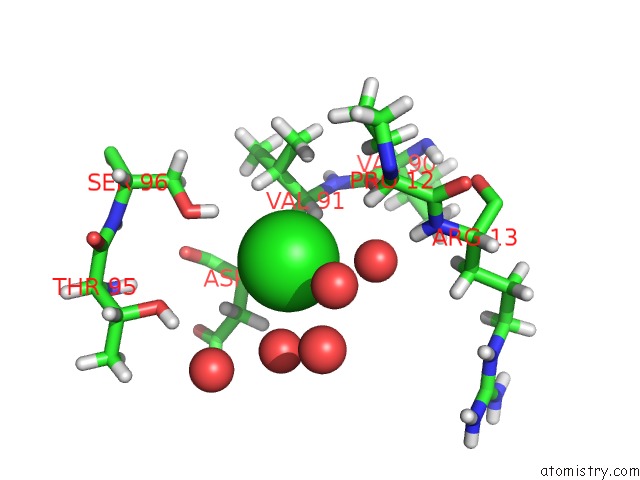
Mono view
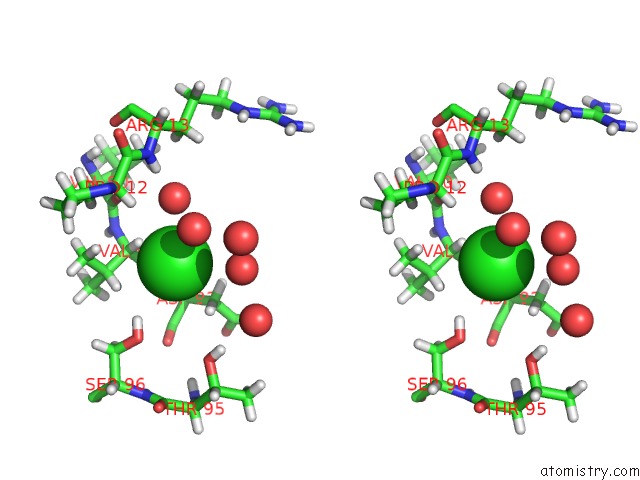
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of the Chemokine Receptor CXCR2 in Complex with the First Pdz Domain of NHERF1 within 5.0Å range:
|
Chlorine binding site 2 out of 4 in 4n6x
Go back to
Chlorine binding site 2 out
of 4 in the Crystal Structure of the Chemokine Receptor CXCR2 in Complex with the First Pdz Domain of NHERF1
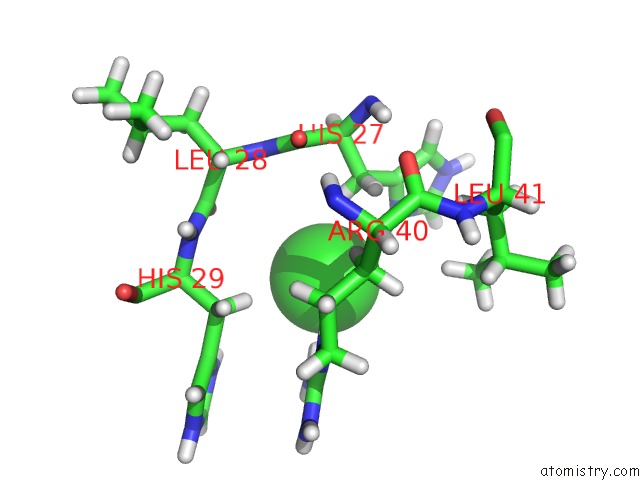
Mono view
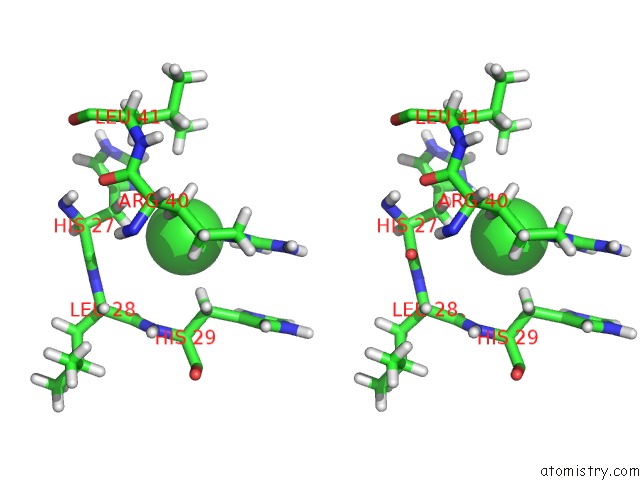
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of the Chemokine Receptor CXCR2 in Complex with the First Pdz Domain of NHERF1 within 5.0Å range:
|
Chlorine binding site 3 out of 4 in 4n6x
Go back to
Chlorine binding site 3 out
of 4 in the Crystal Structure of the Chemokine Receptor CXCR2 in Complex with the First Pdz Domain of NHERF1
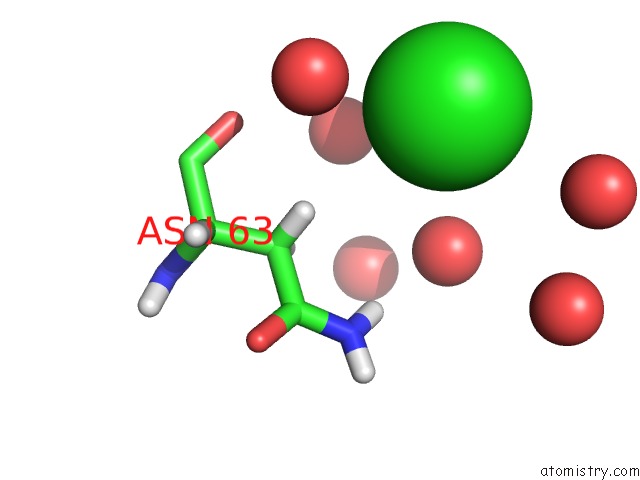
Mono view
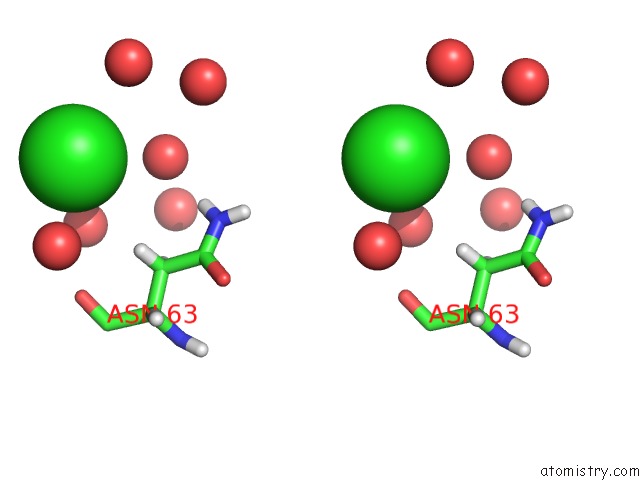
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Crystal Structure of the Chemokine Receptor CXCR2 in Complex with the First Pdz Domain of NHERF1 within 5.0Å range:
|
Chlorine binding site 4 out of 4 in 4n6x
Go back to
Chlorine binding site 4 out
of 4 in the Crystal Structure of the Chemokine Receptor CXCR2 in Complex with the First Pdz Domain of NHERF1
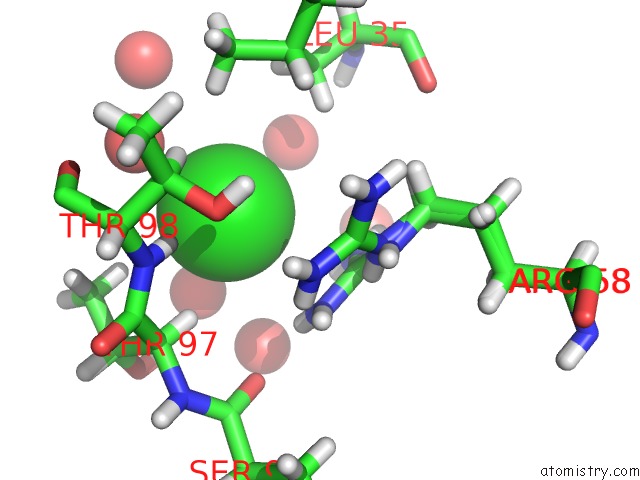
Mono view
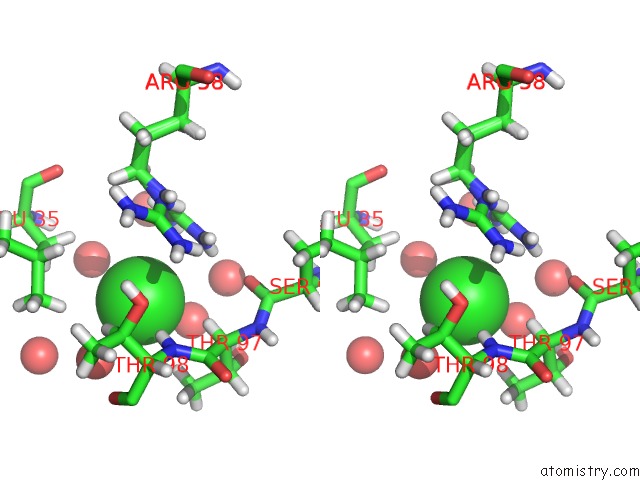
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Crystal Structure of the Chemokine Receptor CXCR2 in Complex with the First Pdz Domain of NHERF1 within 5.0Å range:
|
Reference:
Y.Jiang,
G.Lu,
L.R.Trescott,
Y.Hou,
X.Guan,
S.Wang,
A.Stamenkovich,
J.Brunzelle,
N.Sirinupong,
C.Li,
Z.Yang.
New Conformational State of NHERF1-CXCR2 Signaling Complex Captured By Crystal Lattice Trapping. Plos One V. 8 81904 2013.
ISSN: ESSN 1932-6203
PubMed: 24339979
DOI: 10.1371/JOURNAL.PONE.0081904
Page generated: Sun Jul 21 20:29:28 2024
ISSN: ESSN 1932-6203
PubMed: 24339979
DOI: 10.1371/JOURNAL.PONE.0081904
Last articles
Ca in 5OLPCa in 5OLQ
Ca in 5OL2
Ca in 5OLB
Ca in 5OL7
Ca in 5OFX
Ca in 5ODU
Ca in 5OHT
Ca in 5OK8
Ca in 5OFI