Chlorine »
PDB 4px1-4q63 »
4q3n »
Chlorine in PDB 4q3n: Crystal Structure of Mgs-M5, A Lactate Dehydrogenase Enzyme From A Medee Basin Deep-Sea Metagenome Library
Protein crystallography data
The structure of Crystal Structure of Mgs-M5, A Lactate Dehydrogenase Enzyme From A Medee Basin Deep-Sea Metagenome Library, PDB code: 4q3n
was solved by
P.J.Stogios,
X.Xu,
H.Cui,
M.Alcaide,
M.Ferrer,
A.Savchenko,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 34.31 / 1.97 |
| Space group | I 41 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 92.818, 92.818, 203.804, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 14.5 / 18.4 |
Other elements in 4q3n:
The structure of Crystal Structure of Mgs-M5, A Lactate Dehydrogenase Enzyme From A Medee Basin Deep-Sea Metagenome Library also contains other interesting chemical elements:
| Sodium | (Na) | 1 atom |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of Mgs-M5, A Lactate Dehydrogenase Enzyme From A Medee Basin Deep-Sea Metagenome Library
(pdb code 4q3n). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Crystal Structure of Mgs-M5, A Lactate Dehydrogenase Enzyme From A Medee Basin Deep-Sea Metagenome Library, PDB code: 4q3n:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Crystal Structure of Mgs-M5, A Lactate Dehydrogenase Enzyme From A Medee Basin Deep-Sea Metagenome Library, PDB code: 4q3n:
Jump to Chlorine binding site number: 1; 2;
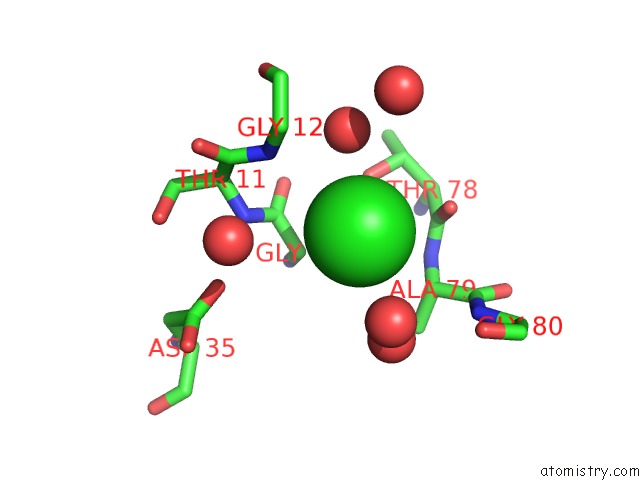
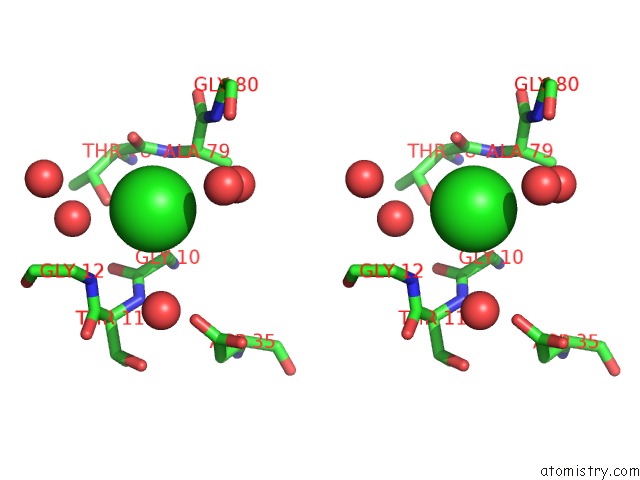
Chlorine binding site 1 out of 2 in 4q3n
Go back to
Chlorine binding site 1 out
of 2 in the Crystal Structure of Mgs-M5, A Lactate Dehydrogenase Enzyme From A Medee Basin Deep-Sea Metagenome Library
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of Mgs-M5, A Lactate Dehydrogenase Enzyme From A Medee Basin Deep-Sea Metagenome Library within 5.0Å range:
|
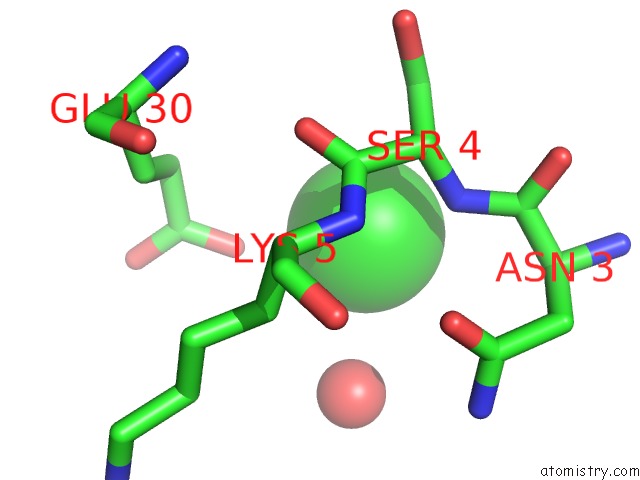
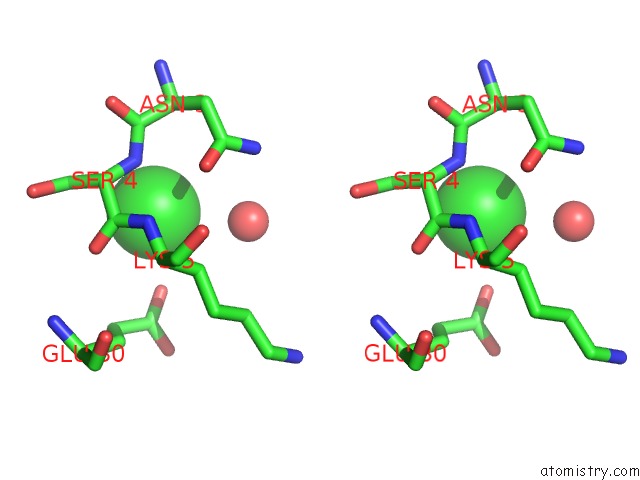
Chlorine binding site 2 out of 2 in 4q3n
Go back to
Chlorine binding site 2 out
of 2 in the Crystal Structure of Mgs-M5, A Lactate Dehydrogenase Enzyme From A Medee Basin Deep-Sea Metagenome Library
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of Mgs-M5, A Lactate Dehydrogenase Enzyme From A Medee Basin Deep-Sea Metagenome Library within 5.0Å range:
|
Reference:
M.Alcaide,
P.J.Stogios,
A.Lafraya,
A.Tchigvintsev,
R.Flick,
R.Bargiela,
T.N.Chernikova,
O.N.Reva,
T.Hai,
C.C.Leggewie,
N.Katzke,
V.La Cono,
R.Matesanz,
M.Jebbar,
K.E.Jaeger,
M.M.Yakimov,
A.F.Yakunin,
P.N.Golyshin,
O.V.Golyshina,
A.Savchenko,
M.Ferrer.
Pressure Adaptation Is Linked to Thermal Adaptation in Salt-Saturated Marine Habitats. Environ Microbiol 2014.
PubMed: 25330254
DOI: 10.1111/1462-2920.12660
Page generated: Fri Jul 11 20:38:59 2025
PubMed: 25330254
DOI: 10.1111/1462-2920.12660
Last articles
F in 4JSVF in 4JTQ
F in 4JSC
F in 4JSJ
F in 4JSM
F in 4JQG
F in 4JSI
F in 4JQ2
F in 4JP4
F in 4JPS