Chlorine »
PDB 4tsz-4u2x »
4u01 »
Chlorine in PDB 4u01: Hcv NS3/4A Serine Protease in Complex with 6570
Protein crystallography data
The structure of Hcv NS3/4A Serine Protease in Complex with 6570, PDB code: 4u01
was solved by
C.C.Parsy,
F.-R.Alexandre,
G.Brandt,
C.Caillet,
D.Chaves,
M.Derock,
D.Gloux,
Y.Griffon,
L.B.Lallos,
F.Leroy,
M.Liuzzi,
A.-G.Loi,
B.Mayes,
L.Moulat,
A.Moussa,
M.Chiara,
V.Roques,
E.Rosinovsky,
M.Seifer,
A.Stewart,
J.Wang,
D.Standring,
D.Surleraux,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 43.23 / 2.80 |
| Space group | C 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 150.034, 174.363, 133.010, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 23.3 / 29 |
Other elements in 4u01:
The structure of Hcv NS3/4A Serine Protease in Complex with 6570 also contains other interesting chemical elements:
| Fluorine | (F) | 9 atoms |
| Zinc | (Zn) | 9 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Hcv NS3/4A Serine Protease in Complex with 6570
(pdb code 4u01). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 4 binding sites of Chlorine where determined in the Hcv NS3/4A Serine Protease in Complex with 6570, PDB code: 4u01:
Jump to Chlorine binding site number: 1; 2; 3; 4;
In total 4 binding sites of Chlorine where determined in the Hcv NS3/4A Serine Protease in Complex with 6570, PDB code: 4u01:
Jump to Chlorine binding site number: 1; 2; 3; 4;
Chlorine binding site 1 out of 4 in 4u01
Go back to
Chlorine binding site 1 out
of 4 in the Hcv NS3/4A Serine Protease in Complex with 6570
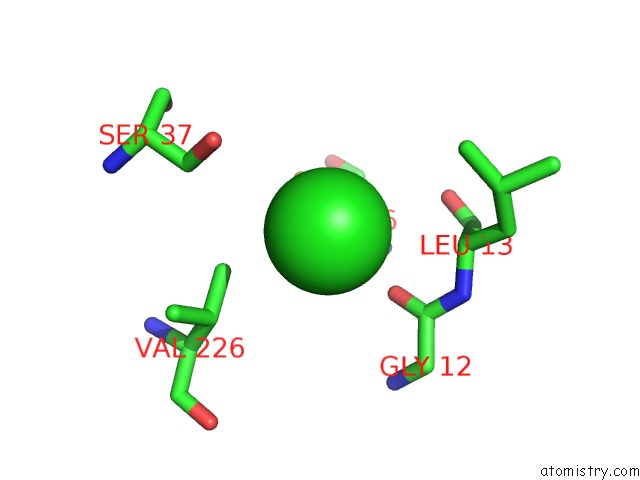
Mono view
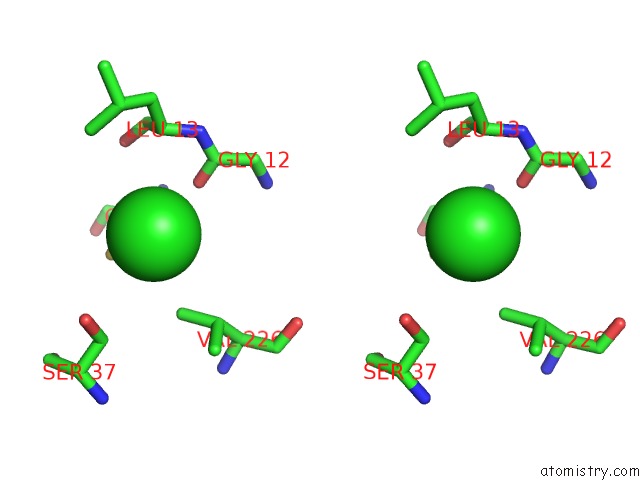
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Hcv NS3/4A Serine Protease in Complex with 6570 within 5.0Å range:
|
Chlorine binding site 2 out of 4 in 4u01
Go back to
Chlorine binding site 2 out
of 4 in the Hcv NS3/4A Serine Protease in Complex with 6570
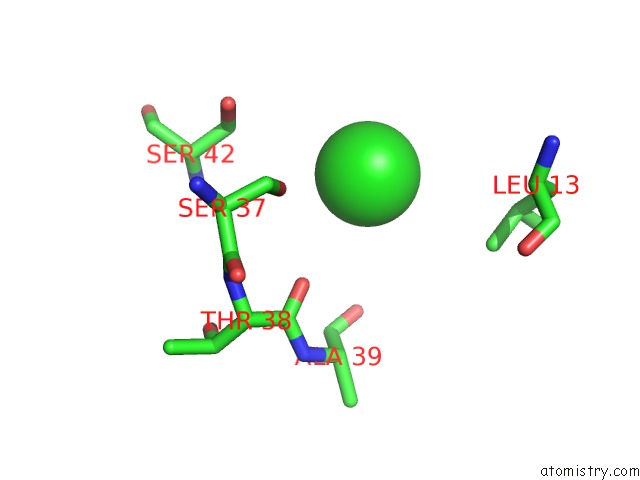
Mono view
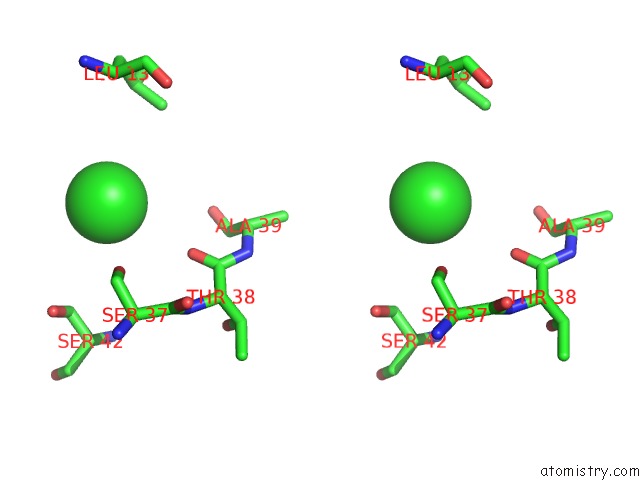
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Hcv NS3/4A Serine Protease in Complex with 6570 within 5.0Å range:
|
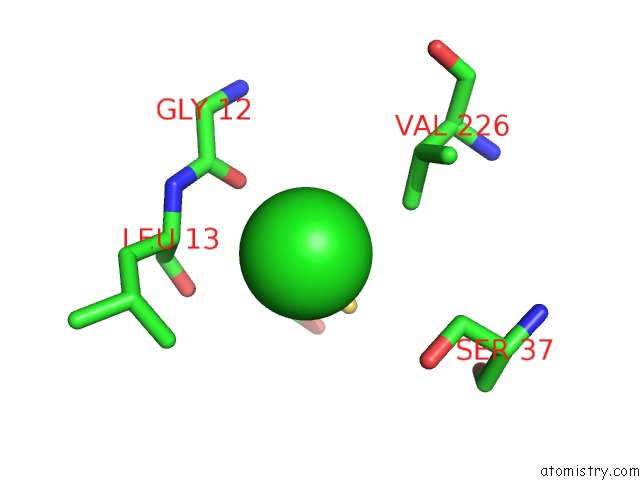
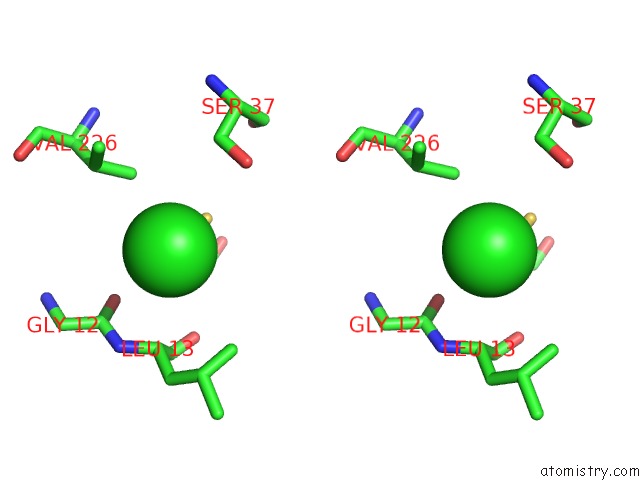
Chlorine binding site 3 out of 4 in 4u01
Go back to
Chlorine binding site 3 out
of 4 in the Hcv NS3/4A Serine Protease in Complex with 6570
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Hcv NS3/4A Serine Protease in Complex with 6570 within 5.0Å range:
|
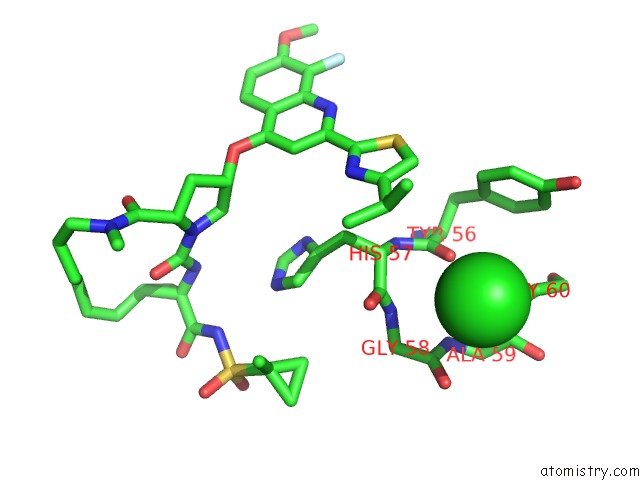
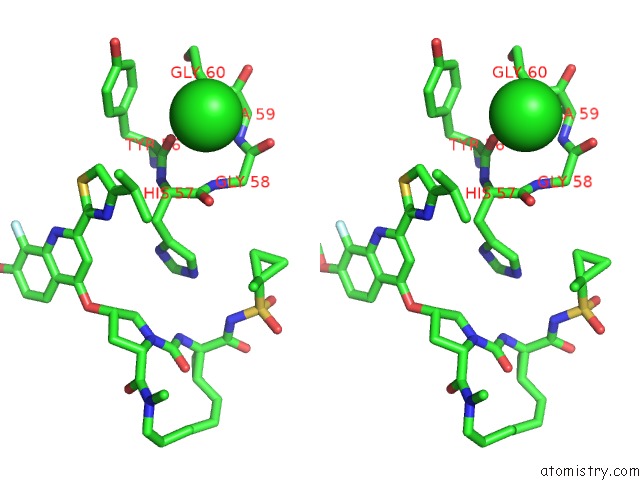
Chlorine binding site 4 out of 4 in 4u01
Go back to
Chlorine binding site 4 out
of 4 in the Hcv NS3/4A Serine Protease in Complex with 6570
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Hcv NS3/4A Serine Protease in Complex with 6570 within 5.0Å range:
|
Reference:
C.C.Parsy,
F.R.Alexandre,
V.Bidau,
F.Bonnaterre,
G.Brandt,
C.Caillet,
S.Cappelle,
D.Chaves,
T.Convard,
M.Derock,
D.Gloux,
Y.Griffon,
L.B.Lallos,
F.Leroy,
M.Liuzzi,
A.G.Loi,
L.Moulat,
M.Chiara,
H.Rahali,
V.Roques,
E.Rosinovsky,
S.Savin,
M.Seifer,
D.Standring,
D.Surleraux.
Discovery and Structural Diversity of the Hepatitis C Virus NS3/4A Serine Protease Inhibitor Series Leading to Clinical Candidate IDX320. Bioorg.Med.Chem.Lett. V. 25 5427 2015.
ISSN: ESSN 1464-3405
PubMed: 26410074
DOI: 10.1016/J.BMCL.2015.09.009
Page generated: Fri Jul 11 21:54:43 2025
ISSN: ESSN 1464-3405
PubMed: 26410074
DOI: 10.1016/J.BMCL.2015.09.009
Last articles
Cl in 7ZSHCl in 7ZSG
Cl in 7ZNM
Cl in 7ZSF
Cl in 7ZQX
Cl in 7ZR4
Cl in 7ZR3
Cl in 7ZQQ
Cl in 7ZQI
Cl in 7ZPG