Chlorine »
PDB 4uvr-4v3k »
4uwy »
Chlorine in PDB 4uwy: FGFR1 Apo Structure
Enzymatic activity of FGFR1 Apo Structure
All present enzymatic activity of FGFR1 Apo Structure:
2.7.10.1;
2.7.10.1;
Protein crystallography data
The structure of FGFR1 Apo Structure, PDB code: 4uwy
was solved by
N.Thiyagarajan,
T.Bunney,
M.Katan,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 27.815 / 2.30 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 211.837, 49.792, 66.054, 90.00, 107.40, 90.00 |
| R / Rfree (%) | 21.14 / 26.66 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the FGFR1 Apo Structure
(pdb code 4uwy). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 3 binding sites of Chlorine where determined in the FGFR1 Apo Structure, PDB code: 4uwy:
Jump to Chlorine binding site number: 1; 2; 3;
In total 3 binding sites of Chlorine where determined in the FGFR1 Apo Structure, PDB code: 4uwy:
Jump to Chlorine binding site number: 1; 2; 3;
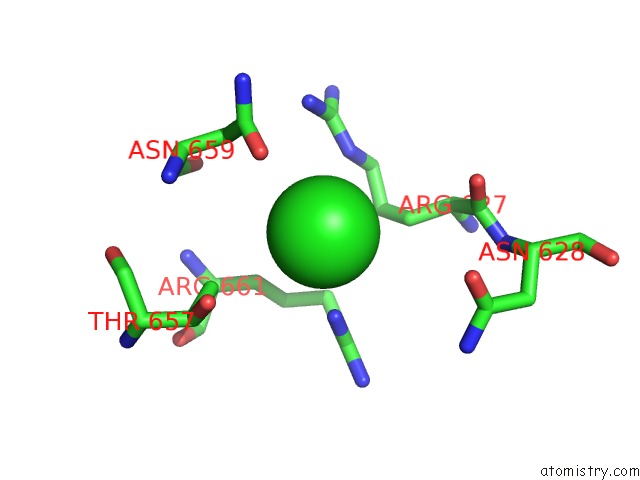
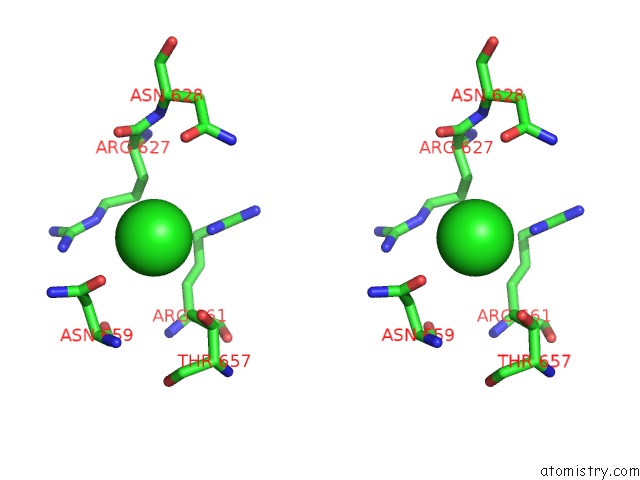
Chlorine binding site 1 out of 3 in 4uwy
Go back to
Chlorine binding site 1 out
of 3 in the FGFR1 Apo Structure
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of FGFR1 Apo Structure within 5.0Å range:
|
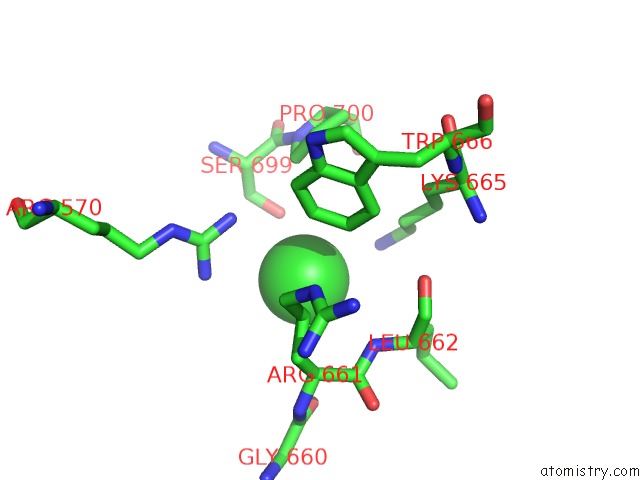
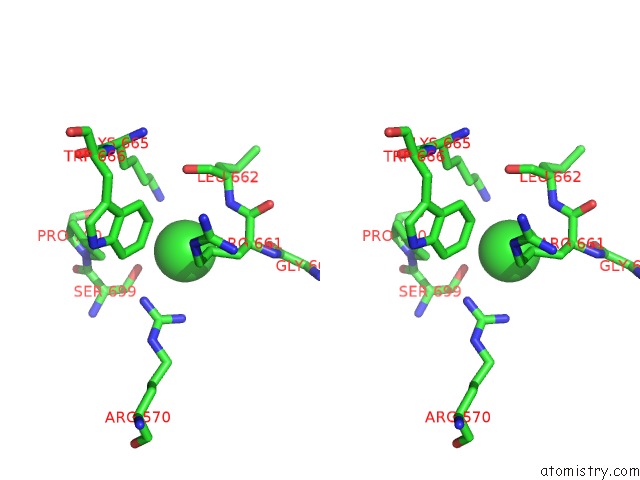
Chlorine binding site 2 out of 3 in 4uwy
Go back to
Chlorine binding site 2 out
of 3 in the FGFR1 Apo Structure
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of FGFR1 Apo Structure within 5.0Å range:
|
Chlorine binding site 3 out of 3 in 4uwy
Go back to
Chlorine binding site 3 out
of 3 in the FGFR1 Apo Structure
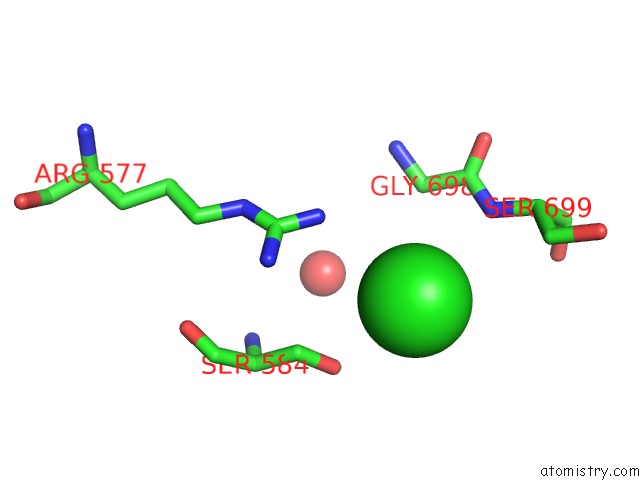
Mono view
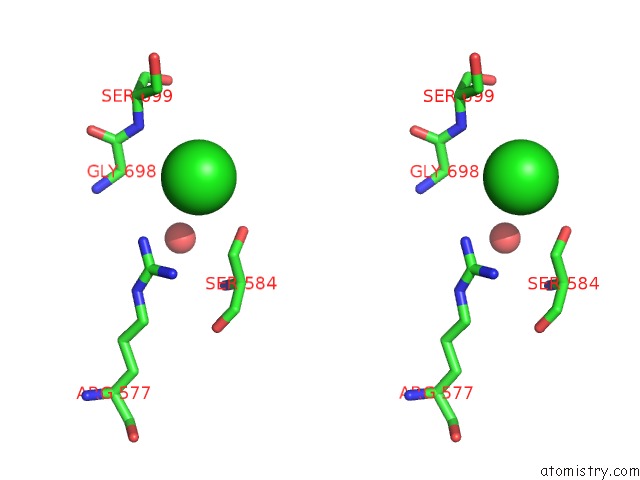
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of FGFR1 Apo Structure within 5.0Å range:
|
Reference:
T.Bunney,
S.Wan,
N.Thiyagarajan,
L.Sutto,
S.V.Williams,
P.Ashford,
H.Koss,
M.A.Knowles,
F.L.Gervasio,
P.V.Coveney,
M.Katan.
The Effect of Mutations on Drug Sensitivity and Kinase Activity of Fibroblast Growth Factor Receptors: A Combined Experimental and Theoretical Study Ebiomedicine 2015.
ISSN: ESSN 2352-3964
DOI: 10.1016/J.EBIOM.2015.02.009
Page generated: Fri Jul 11 22:17:54 2025
ISSN: ESSN 2352-3964
DOI: 10.1016/J.EBIOM.2015.02.009
Last articles
F in 7Q2YF in 7Q3B
F in 7Q2X
F in 7PVK
F in 7Q2J
F in 7Q01
F in 7PZX
F in 7PZW
F in 7PZV
F in 7PZU