Chlorine »
PDB 4z6v-4zg7 »
4z84 »
Chlorine in PDB 4z84: PKAB3 in Complex with Pyrrolidine Inhibitor 34A
Enzymatic activity of PKAB3 in Complex with Pyrrolidine Inhibitor 34A
All present enzymatic activity of PKAB3 in Complex with Pyrrolidine Inhibitor 34A:
2.7.11.11;
2.7.11.11;
Protein crystallography data
The structure of PKAB3 in Complex with Pyrrolidine Inhibitor 34A, PDB code: 4z84
was solved by
B.A.Lund,
K.A.Alam,
R.A.Engh,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 19.61 / 1.55 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 61.852, 78.431, 79.506, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20.8 / 23.4 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the PKAB3 in Complex with Pyrrolidine Inhibitor 34A
(pdb code 4z84). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the PKAB3 in Complex with Pyrrolidine Inhibitor 34A, PDB code: 4z84:
In total only one binding site of Chlorine was determined in the PKAB3 in Complex with Pyrrolidine Inhibitor 34A, PDB code: 4z84:
Chlorine binding site 1 out of 1 in 4z84
Go back to
Chlorine binding site 1 out
of 1 in the PKAB3 in Complex with Pyrrolidine Inhibitor 34A
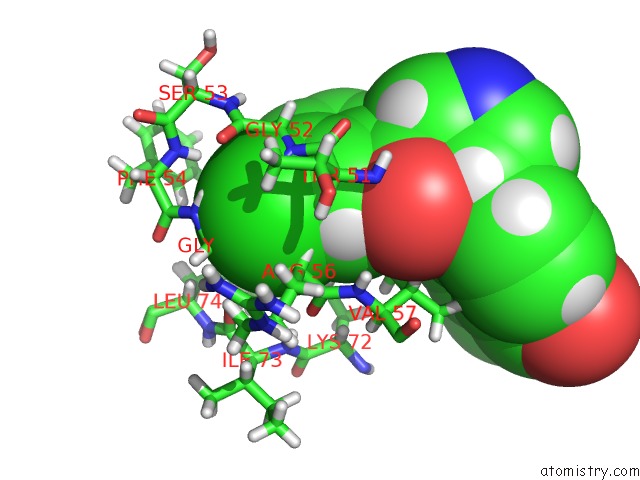
Mono view
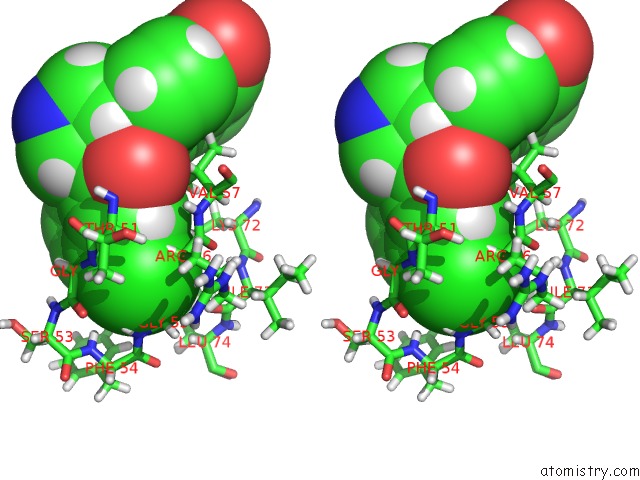
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of PKAB3 in Complex with Pyrrolidine Inhibitor 34A within 5.0Å range:
|
Reference:
B.S.Lauber,
L.A.Hardegger,
A.K.Asraful,
B.A.Lund,
O.Dumele,
M.Harder,
B.Kuhn,
R.A.Engh,
F.Diederich.
Addressing the Glycine-Rich Loop of Protein Kinases By A Multi-Facetted Interaction Network: Inhibition of Pka and A Pkb Mimic. Chemistry V. 22 211 2016.
ISSN: ISSN 0947-6539
PubMed: 26578105
DOI: 10.1002/CHEM.201503552
Page generated: Fri Jul 11 23:38:12 2025
ISSN: ISSN 0947-6539
PubMed: 26578105
DOI: 10.1002/CHEM.201503552
Last articles
Cl in 7YRACl in 7YR3
Cl in 7YQ7
Cl in 7YQ2
Cl in 7YQN
Cl in 7YR2
Cl in 7YQ9
Cl in 7YPZ
Cl in 7YNV
Cl in 7YNU