Chlorine »
PDB 5hqa-5i1f »
5i0d »
Chlorine in PDB 5i0d: Cycloalternan-Forming Enzyme From Listeria Monocytogenes in Complex with Cycloalternan
Protein crystallography data
The structure of Cycloalternan-Forming Enzyme From Listeria Monocytogenes in Complex with Cycloalternan, PDB code: 5i0d
was solved by
S.H.Light,
G.Minasov,
W.F.Anderson,
Center For Structural Genomics Ofinfectious Diseases (Csgid),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 30.00 / 1.77 |
| Space group | P 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 74.753, 101.233, 166.391, 90.00, 101.02, 90.00 |
| R / Rfree (%) | 14.4 / 17.1 |
Other elements in 5i0d:
The structure of Cycloalternan-Forming Enzyme From Listeria Monocytogenes in Complex with Cycloalternan also contains other interesting chemical elements:
| Magnesium | (Mg) | 8 atoms |
| Calcium | (Ca) | 2 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Cycloalternan-Forming Enzyme From Listeria Monocytogenes in Complex with Cycloalternan
(pdb code 5i0d). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 7 binding sites of Chlorine where determined in the Cycloalternan-Forming Enzyme From Listeria Monocytogenes in Complex with Cycloalternan, PDB code: 5i0d:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7;
In total 7 binding sites of Chlorine where determined in the Cycloalternan-Forming Enzyme From Listeria Monocytogenes in Complex with Cycloalternan, PDB code: 5i0d:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7;
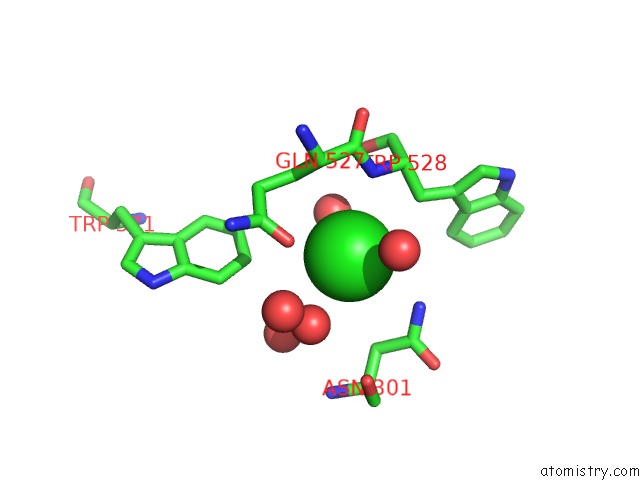
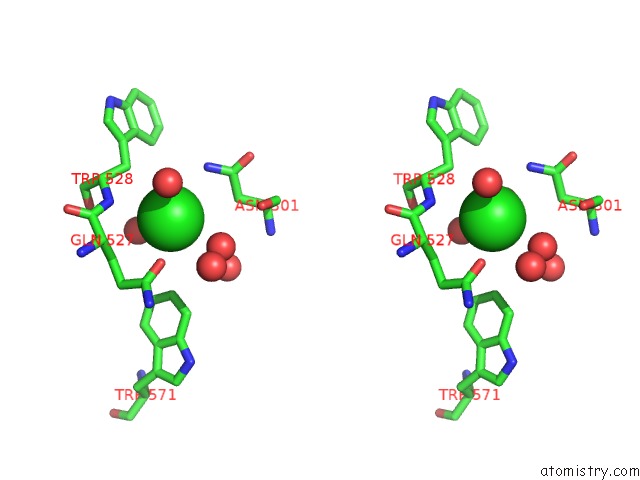
Chlorine binding site 1 out of 7 in 5i0d
Go back to
Chlorine binding site 1 out
of 7 in the Cycloalternan-Forming Enzyme From Listeria Monocytogenes in Complex with Cycloalternan
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Cycloalternan-Forming Enzyme From Listeria Monocytogenes in Complex with Cycloalternan within 5.0Å range:
|
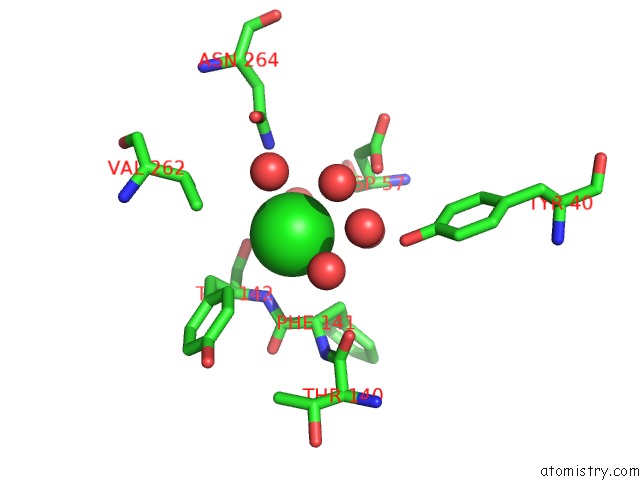
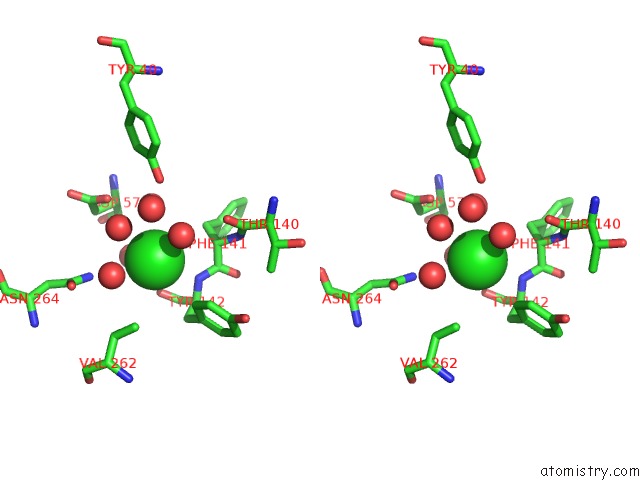
Chlorine binding site 2 out of 7 in 5i0d
Go back to
Chlorine binding site 2 out
of 7 in the Cycloalternan-Forming Enzyme From Listeria Monocytogenes in Complex with Cycloalternan
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Cycloalternan-Forming Enzyme From Listeria Monocytogenes in Complex with Cycloalternan within 5.0Å range:
|
Chlorine binding site 3 out of 7 in 5i0d
Go back to
Chlorine binding site 3 out
of 7 in the Cycloalternan-Forming Enzyme From Listeria Monocytogenes in Complex with Cycloalternan
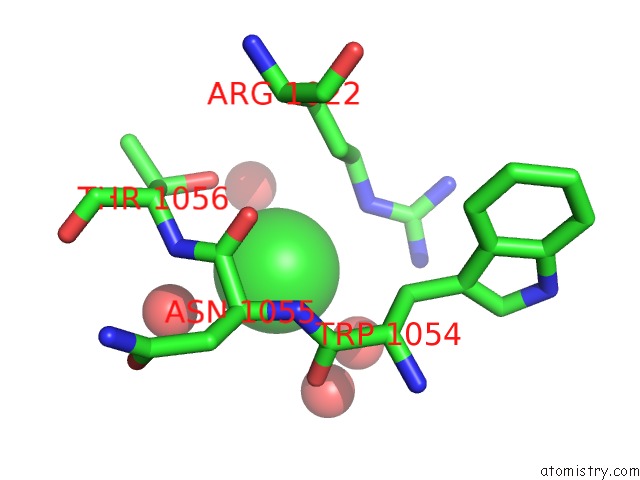
Mono view
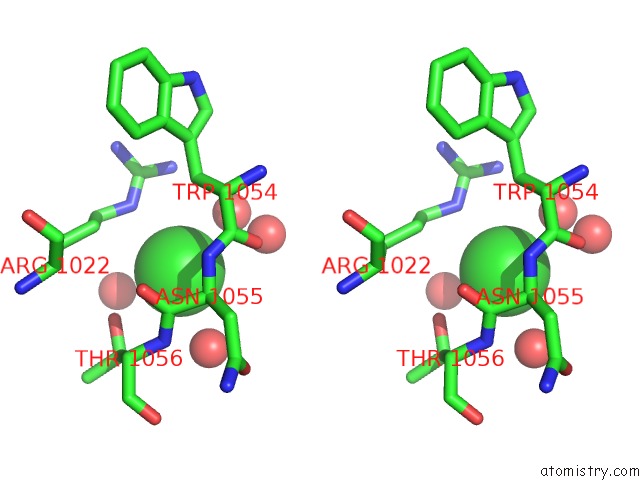
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Cycloalternan-Forming Enzyme From Listeria Monocytogenes in Complex with Cycloalternan within 5.0Å range:
|
Chlorine binding site 4 out of 7 in 5i0d
Go back to
Chlorine binding site 4 out
of 7 in the Cycloalternan-Forming Enzyme From Listeria Monocytogenes in Complex with Cycloalternan
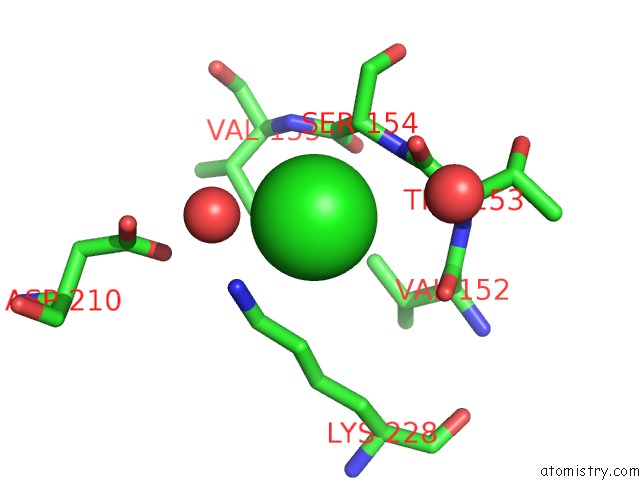
Mono view
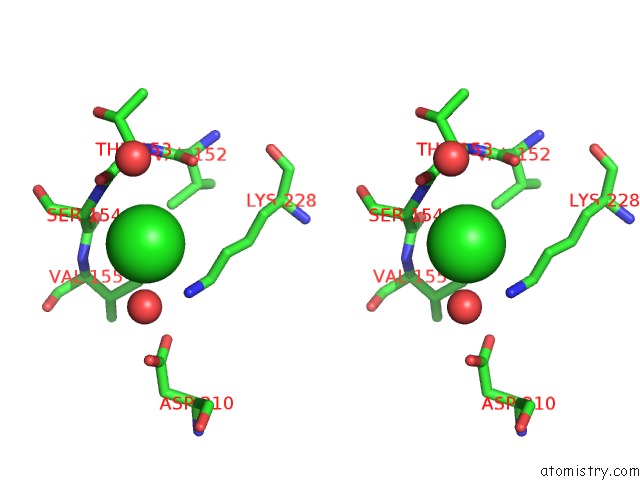
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Cycloalternan-Forming Enzyme From Listeria Monocytogenes in Complex with Cycloalternan within 5.0Å range:
|
Chlorine binding site 5 out of 7 in 5i0d
Go back to
Chlorine binding site 5 out
of 7 in the Cycloalternan-Forming Enzyme From Listeria Monocytogenes in Complex with Cycloalternan
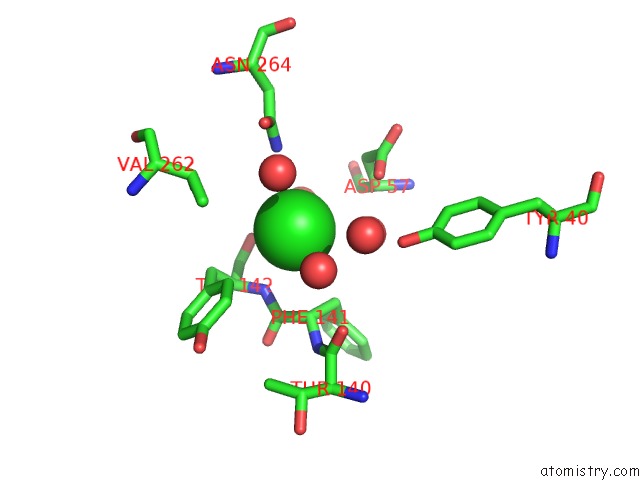
Mono view
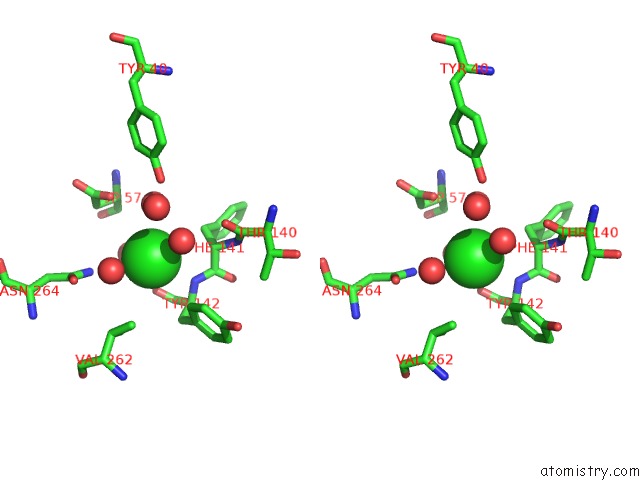
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Cycloalternan-Forming Enzyme From Listeria Monocytogenes in Complex with Cycloalternan within 5.0Å range:
|
Chlorine binding site 6 out of 7 in 5i0d
Go back to
Chlorine binding site 6 out
of 7 in the Cycloalternan-Forming Enzyme From Listeria Monocytogenes in Complex with Cycloalternan
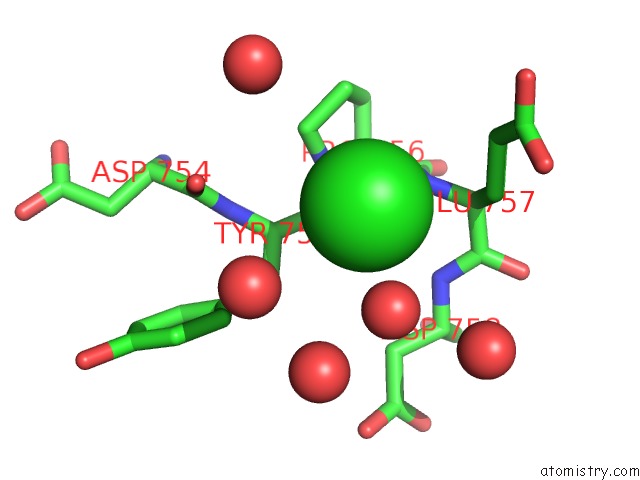
Mono view
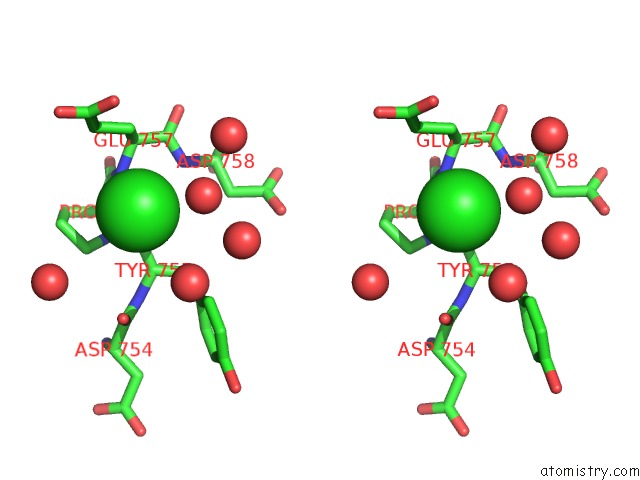
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 6 of Cycloalternan-Forming Enzyme From Listeria Monocytogenes in Complex with Cycloalternan within 5.0Å range:
|
Chlorine binding site 7 out of 7 in 5i0d
Go back to
Chlorine binding site 7 out
of 7 in the Cycloalternan-Forming Enzyme From Listeria Monocytogenes in Complex with Cycloalternan
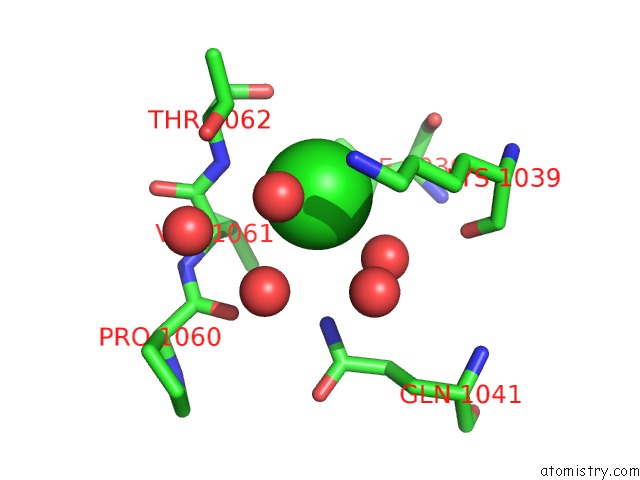
Mono view
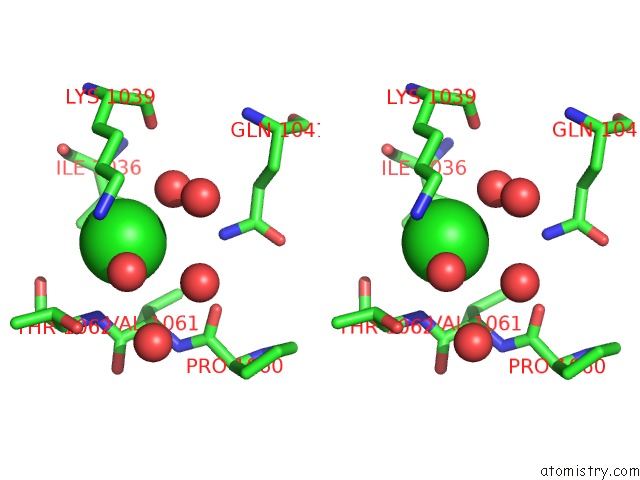
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 7 of Cycloalternan-Forming Enzyme From Listeria Monocytogenes in Complex with Cycloalternan within 5.0Å range:
|
Reference:
S.H.Light,
L.A.Cahoon,
K.V.Mahasenan,
M.Lee,
B.Boggess,
A.S.Halavaty,
S.Mobashery,
N.E.Freitag,
W.F.Anderson.
Transferase Versus Hydrolase: the Role of Conformational Flexibility in Reaction Specificity. Structure V. 25 295 2017.
ISSN: ISSN 1878-4186
PubMed: 28089449
DOI: 10.1016/J.STR.2016.12.007
Page generated: Sat Jul 12 02:56:25 2025
ISSN: ISSN 1878-4186
PubMed: 28089449
DOI: 10.1016/J.STR.2016.12.007
Last articles
F in 4KN6F in 4KKL
F in 4KMN
F in 4KMP
F in 4KKX
F in 4KK8
F in 4KJQ
F in 4KHM
F in 4KE5
F in 4KHR