Chlorine »
PDB 5mzf-5n6t »
5mzv »
Chlorine in PDB 5mzv: Il-23:Il-23R:NB22E11 Complex
Protein crystallography data
The structure of Il-23:Il-23R:NB22E11 Complex, PDB code: 5mzv
was solved by
Y.Bloch,
S.N.Savvides,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 76.85 / 2.80 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 67.219, 112.190, 109.870, 90.00, 106.24, 90.00 |
| R / Rfree (%) | 21.2 / 25 |
Other elements in 5mzv:
The structure of Il-23:Il-23R:NB22E11 Complex also contains other interesting chemical elements:
| Sodium | (Na) | 1 atom |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Il-23:Il-23R:NB22E11 Complex
(pdb code 5mzv). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 3 binding sites of Chlorine where determined in the Il-23:Il-23R:NB22E11 Complex, PDB code: 5mzv:
Jump to Chlorine binding site number: 1; 2; 3;
In total 3 binding sites of Chlorine where determined in the Il-23:Il-23R:NB22E11 Complex, PDB code: 5mzv:
Jump to Chlorine binding site number: 1; 2; 3;
Chlorine binding site 1 out of 3 in 5mzv
Go back to
Chlorine binding site 1 out
of 3 in the Il-23:Il-23R:NB22E11 Complex
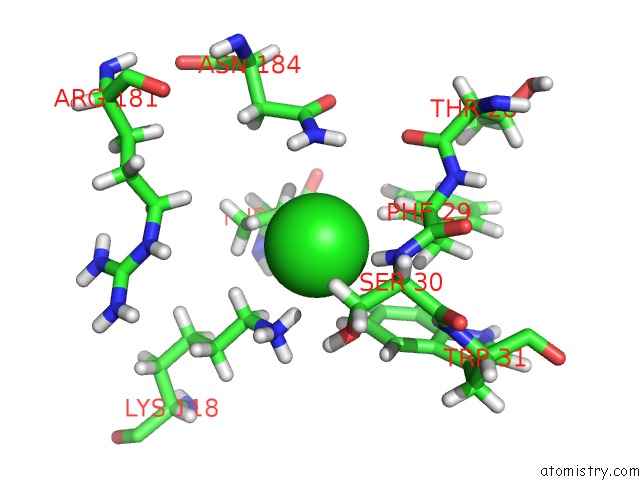
Mono view
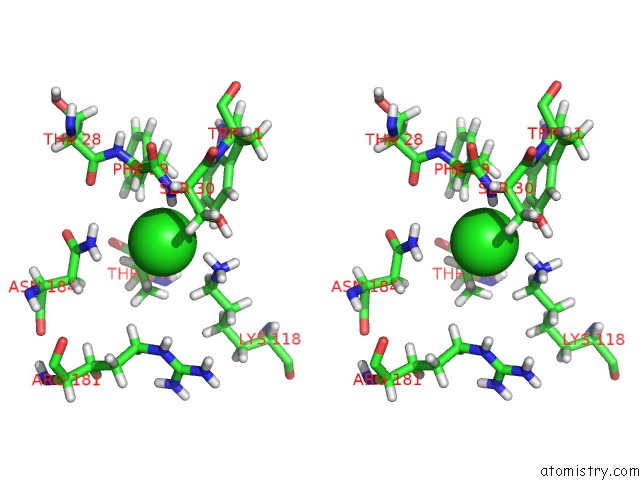
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Il-23:Il-23R:NB22E11 Complex within 5.0Å range:
|
Chlorine binding site 2 out of 3 in 5mzv
Go back to
Chlorine binding site 2 out
of 3 in the Il-23:Il-23R:NB22E11 Complex
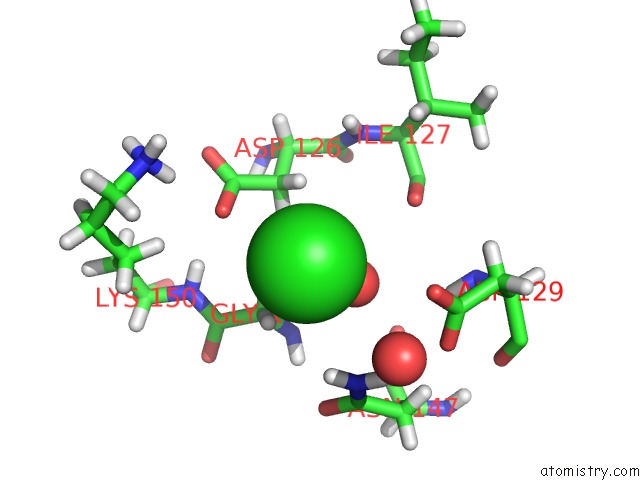
Mono view
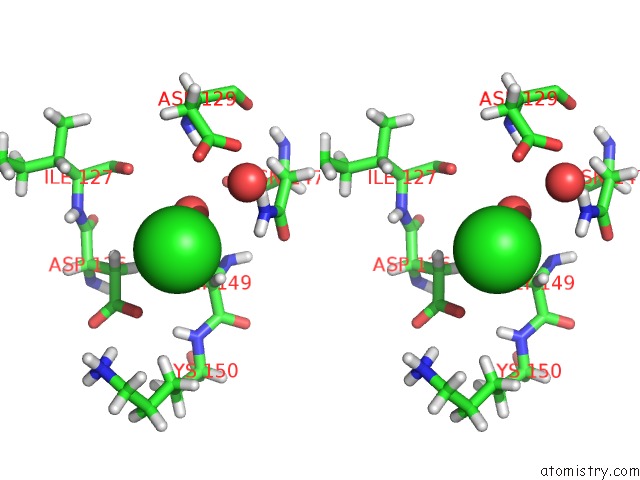
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Il-23:Il-23R:NB22E11 Complex within 5.0Å range:
|
Chlorine binding site 3 out of 3 in 5mzv
Go back to
Chlorine binding site 3 out
of 3 in the Il-23:Il-23R:NB22E11 Complex
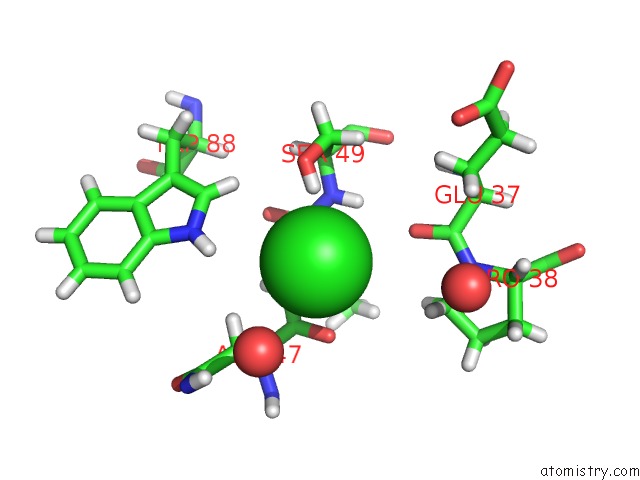
Mono view
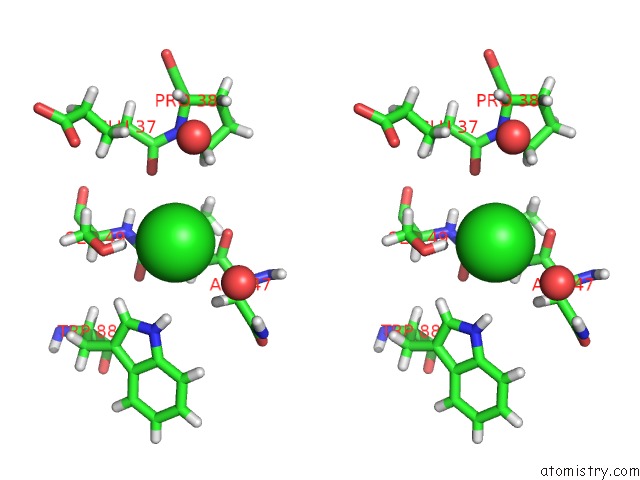
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Il-23:Il-23R:NB22E11 Complex within 5.0Å range:
|
Reference:
Y.Bloch,
L.Bouchareychas,
R.Merceron,
K.Skladanowska,
L.Van Den Bossche,
S.Detry,
S.Govindarajan,
D.Elewaut,
F.Haerynck,
M.Dullaers,
I.E.Adamopoulos,
S.N.Savvides.
Structural Activation of Pro-Inflammatory Human Cytokine Il-23 By Cognate Il-23 Receptor Enables Recruitment of the Shared Receptor Il-12R Beta 1. Immunity V. 48 45 2018.
ISSN: ISSN 1097-4180
PubMed: 29287995
DOI: 10.1016/J.IMMUNI.2017.12.008
Page generated: Fri Jul 26 13:05:31 2024
ISSN: ISSN 1097-4180
PubMed: 29287995
DOI: 10.1016/J.IMMUNI.2017.12.008
Last articles
Cl in 3ZM4Cl in 3ZLX
Cl in 3ZL1
Cl in 3ZKJ
Cl in 3ZJK
Cl in 3ZKX
Cl in 3ZKQ
Cl in 3ZJG
Cl in 3ZJE
Cl in 3ZHC