Chlorine »
PDB 5mzf-5n6t »
5n58 »
Chlorine in PDB 5n58: Di-Zinc Vim-5 Metallo-Beta-Lactamase in Complex with (1-Chloro-4- Hydroxyisoquinoline-3-Carbonyl)-D-Tryptophan (Compound 1)
Protein crystallography data
The structure of Di-Zinc Vim-5 Metallo-Beta-Lactamase in Complex with (1-Chloro-4- Hydroxyisoquinoline-3-Carbonyl)-D-Tryptophan (Compound 1), PDB code: 5n58
was solved by
G.-B.Li,
J.Brem,
M.A.Mcdonough,
C.J.Schofield,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 62.12 / 1.96 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 54.471, 81.540, 95.892, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 18.7 / 23.7 |
Other elements in 5n58:
The structure of Di-Zinc Vim-5 Metallo-Beta-Lactamase in Complex with (1-Chloro-4- Hydroxyisoquinoline-3-Carbonyl)-D-Tryptophan (Compound 1) also contains other interesting chemical elements:
| Zinc | (Zn) | 3 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Di-Zinc Vim-5 Metallo-Beta-Lactamase in Complex with (1-Chloro-4- Hydroxyisoquinoline-3-Carbonyl)-D-Tryptophan (Compound 1)
(pdb code 5n58). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Di-Zinc Vim-5 Metallo-Beta-Lactamase in Complex with (1-Chloro-4- Hydroxyisoquinoline-3-Carbonyl)-D-Tryptophan (Compound 1), PDB code: 5n58:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Di-Zinc Vim-5 Metallo-Beta-Lactamase in Complex with (1-Chloro-4- Hydroxyisoquinoline-3-Carbonyl)-D-Tryptophan (Compound 1), PDB code: 5n58:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 5n58
Go back to
Chlorine binding site 1 out
of 2 in the Di-Zinc Vim-5 Metallo-Beta-Lactamase in Complex with (1-Chloro-4- Hydroxyisoquinoline-3-Carbonyl)-D-Tryptophan (Compound 1)
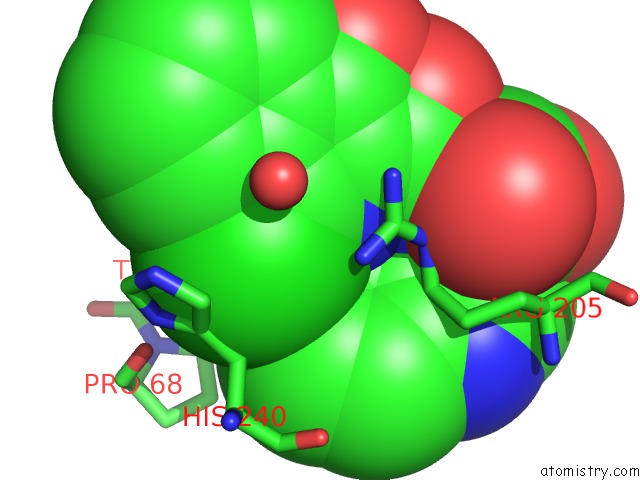
Mono view
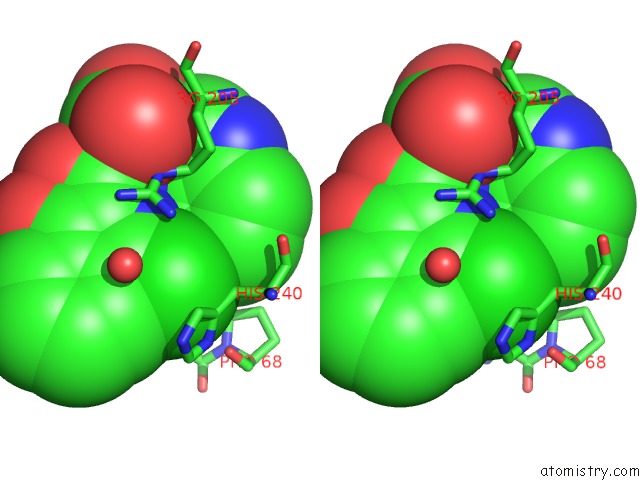
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Di-Zinc Vim-5 Metallo-Beta-Lactamase in Complex with (1-Chloro-4- Hydroxyisoquinoline-3-Carbonyl)-D-Tryptophan (Compound 1) within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 5n58
Go back to
Chlorine binding site 2 out
of 2 in the Di-Zinc Vim-5 Metallo-Beta-Lactamase in Complex with (1-Chloro-4- Hydroxyisoquinoline-3-Carbonyl)-D-Tryptophan (Compound 1)
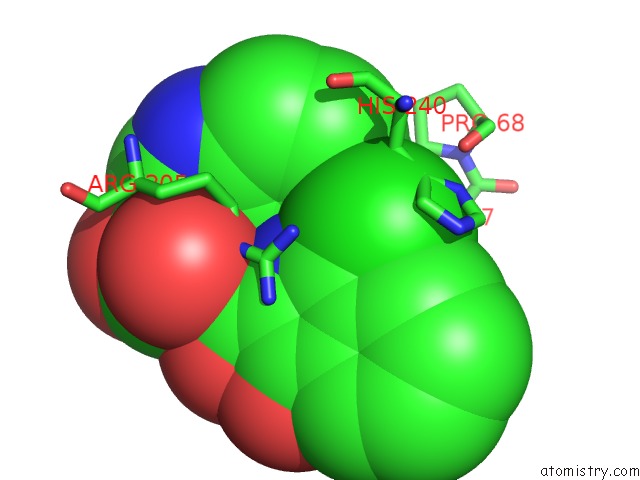
Mono view
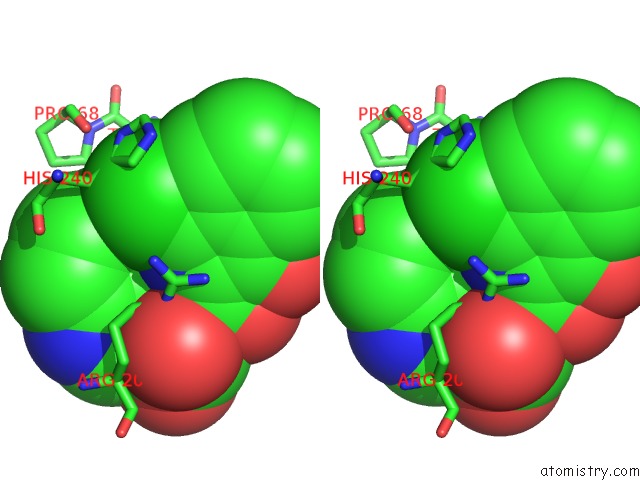
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Di-Zinc Vim-5 Metallo-Beta-Lactamase in Complex with (1-Chloro-4- Hydroxyisoquinoline-3-Carbonyl)-D-Tryptophan (Compound 1) within 5.0Å range:
|
Reference:
G.B.Li,
J.Brem,
R.Lesniak,
M.I.Abboud,
C.T.Lohans,
I.J.Clifton,
S.Y.Yang,
J.C.Jimenez-Castellanos,
M.B.Avison,
J.Spencer,
M.A.Mcdonough,
C.J.Schofield.
Crystallographic Analyses of Isoquinoline Complexes Reveal A New Mode of Metallo-Beta-Lactamase Inhibition. Chem. Commun. (Camb.) V. 53 5806 2017.
ISSN: ESSN 1364-548X
PubMed: 28470248
DOI: 10.1039/C7CC02394D
Page generated: Sat Jul 12 05:52:09 2025
ISSN: ESSN 1364-548X
PubMed: 28470248
DOI: 10.1039/C7CC02394D
Last articles
F in 4IVOF in 4IV4
F in 4IVM
F in 4IV2
F in 4IUI
F in 4IN4
F in 4IU7
F in 4ITI
F in 4IUE
F in 4IRU