Chlorine »
PDB 5ofs-5on9 »
5olu »
Chlorine in PDB 5olu: The Crystal Structure of A Highly Thermostable Carboxyl Esterase From Bacillus Coagulans in Complex with Glycerol
Protein crystallography data
The structure of The Crystal Structure of A Highly Thermostable Carboxyl Esterase From Bacillus Coagulans in Complex with Glycerol, PDB code: 5olu
was solved by
L.J.Gourlay,
C.Nakhnoukh,
M.Bolognesi,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 39.73 / 1.80 |
| Space group | P 63 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 137.644, 137.644, 83.636, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 14.8 / 16.9 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the The Crystal Structure of A Highly Thermostable Carboxyl Esterase From Bacillus Coagulans in Complex with Glycerol
(pdb code 5olu). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 8 binding sites of Chlorine where determined in the The Crystal Structure of A Highly Thermostable Carboxyl Esterase From Bacillus Coagulans in Complex with Glycerol, PDB code: 5olu:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
In total 8 binding sites of Chlorine where determined in the The Crystal Structure of A Highly Thermostable Carboxyl Esterase From Bacillus Coagulans in Complex with Glycerol, PDB code: 5olu:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
Chlorine binding site 1 out of 8 in 5olu
Go back to
Chlorine binding site 1 out
of 8 in the The Crystal Structure of A Highly Thermostable Carboxyl Esterase From Bacillus Coagulans in Complex with Glycerol

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of The Crystal Structure of A Highly Thermostable Carboxyl Esterase From Bacillus Coagulans in Complex with Glycerol within 5.0Å range:
|
Chlorine binding site 2 out of 8 in 5olu
Go back to
Chlorine binding site 2 out
of 8 in the The Crystal Structure of A Highly Thermostable Carboxyl Esterase From Bacillus Coagulans in Complex with Glycerol

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of The Crystal Structure of A Highly Thermostable Carboxyl Esterase From Bacillus Coagulans in Complex with Glycerol within 5.0Å range:
|
Chlorine binding site 3 out of 8 in 5olu
Go back to
Chlorine binding site 3 out
of 8 in the The Crystal Structure of A Highly Thermostable Carboxyl Esterase From Bacillus Coagulans in Complex with Glycerol

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of The Crystal Structure of A Highly Thermostable Carboxyl Esterase From Bacillus Coagulans in Complex with Glycerol within 5.0Å range:
|
Chlorine binding site 4 out of 8 in 5olu
Go back to
Chlorine binding site 4 out
of 8 in the The Crystal Structure of A Highly Thermostable Carboxyl Esterase From Bacillus Coagulans in Complex with Glycerol
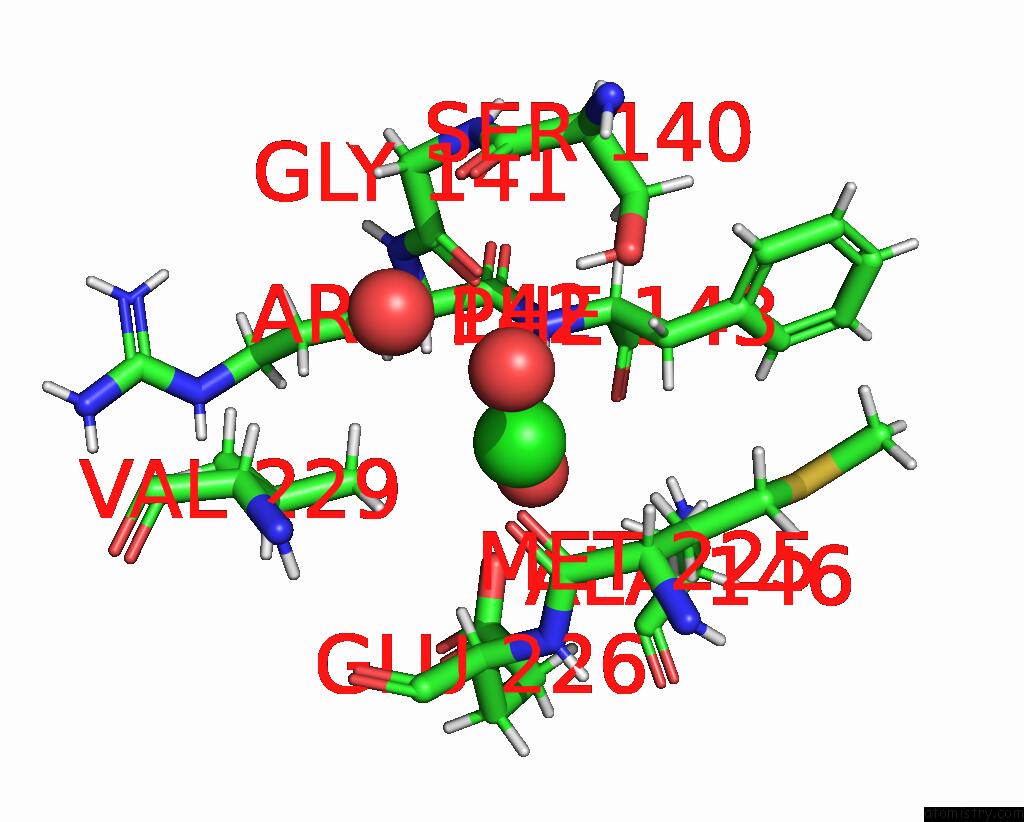
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of The Crystal Structure of A Highly Thermostable Carboxyl Esterase From Bacillus Coagulans in Complex with Glycerol within 5.0Å range:
|
Chlorine binding site 5 out of 8 in 5olu
Go back to
Chlorine binding site 5 out
of 8 in the The Crystal Structure of A Highly Thermostable Carboxyl Esterase From Bacillus Coagulans in Complex with Glycerol

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of The Crystal Structure of A Highly Thermostable Carboxyl Esterase From Bacillus Coagulans in Complex with Glycerol within 5.0Å range:
|
Chlorine binding site 6 out of 8 in 5olu
Go back to
Chlorine binding site 6 out
of 8 in the The Crystal Structure of A Highly Thermostable Carboxyl Esterase From Bacillus Coagulans in Complex with Glycerol

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 6 of The Crystal Structure of A Highly Thermostable Carboxyl Esterase From Bacillus Coagulans in Complex with Glycerol within 5.0Å range:
|
Chlorine binding site 7 out of 8 in 5olu
Go back to
Chlorine binding site 7 out
of 8 in the The Crystal Structure of A Highly Thermostable Carboxyl Esterase From Bacillus Coagulans in Complex with Glycerol

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 7 of The Crystal Structure of A Highly Thermostable Carboxyl Esterase From Bacillus Coagulans in Complex with Glycerol within 5.0Å range:
|
Chlorine binding site 8 out of 8 in 5olu
Go back to
Chlorine binding site 8 out
of 8 in the The Crystal Structure of A Highly Thermostable Carboxyl Esterase From Bacillus Coagulans in Complex with Glycerol

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 8 of The Crystal Structure of A Highly Thermostable Carboxyl Esterase From Bacillus Coagulans in Complex with Glycerol within 5.0Å range:
|
Reference:
V.De Vitis,
C.Nakhnoukh,
A.Pinto,
M.L.Contente,
A.Barbiroli,
M.Milani,
M.Bolognesi,
F.Molinari,
L.J.Gourlay,
D.Romano.
A Stereospecific Carboxyl Esterase From Bacillus Coagulans Hosting Nonlipase Activity Within A Lipase-Like Fold. Febs J. V. 285 903 2018.
ISSN: ISSN 1742-4658
PubMed: 29278448
DOI: 10.1111/FEBS.14368
Page generated: Fri Jul 26 14:33:05 2024
ISSN: ISSN 1742-4658
PubMed: 29278448
DOI: 10.1111/FEBS.14368
Last articles
Cl in 2VOICl in 2VOB
Cl in 2VO6
Cl in 2VO7
Cl in 2VOF
Cl in 2VO5
Cl in 2VMF
Cl in 2VNT
Cl in 2VO3
Cl in 2VO0