Chlorine »
PDB 5qjn-5qtf »
5qqs »
Chlorine in PDB 5qqs: Pandda Analysis Group Deposition -- Crystal Structure of Human ALAS2A in Complex with Z275151340
Enzymatic activity of Pandda Analysis Group Deposition -- Crystal Structure of Human ALAS2A in Complex with Z275151340
All present enzymatic activity of Pandda Analysis Group Deposition -- Crystal Structure of Human ALAS2A in Complex with Z275151340:
2.3.1.37;
2.3.1.37;
Protein crystallography data
The structure of Pandda Analysis Group Deposition -- Crystal Structure of Human ALAS2A in Complex with Z275151340, PDB code: 5qqs
was solved by
G.A.Bezerra,
W.Foster,
H.Bailey,
L.Shrestha,
T.Krojer,
R.Talon,
J.Brandao-Neto,
A.Douangamath,
B.B.Nicola,
F.Von Delft,
C.H.Arrowsmith,
A.Edwards,
C.Bountra,
P.E.Brennan,
W.W.Yue,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 71.48 / 1.85 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 125.949, 108.022, 75.685, 90.00, 109.42, 90.00 |
| R / Rfree (%) | 20.4 / 23.8 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Pandda Analysis Group Deposition -- Crystal Structure of Human ALAS2A in Complex with Z275151340
(pdb code 5qqs). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the Pandda Analysis Group Deposition -- Crystal Structure of Human ALAS2A in Complex with Z275151340, PDB code: 5qqs:
In total only one binding site of Chlorine was determined in the Pandda Analysis Group Deposition -- Crystal Structure of Human ALAS2A in Complex with Z275151340, PDB code: 5qqs:
Chlorine binding site 1 out of 1 in 5qqs
Go back to
Chlorine binding site 1 out
of 1 in the Pandda Analysis Group Deposition -- Crystal Structure of Human ALAS2A in Complex with Z275151340
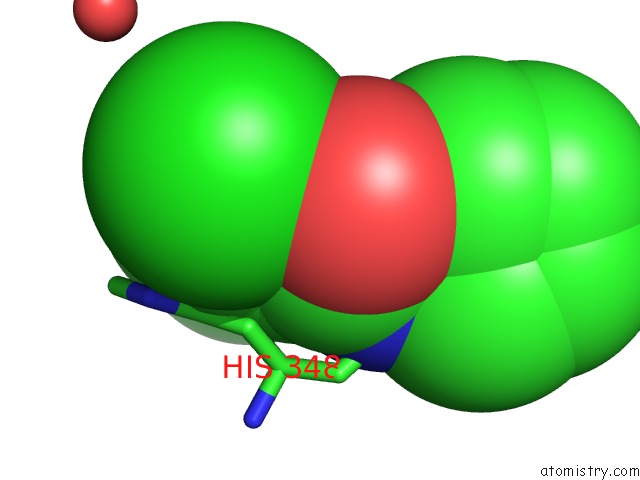
Mono view
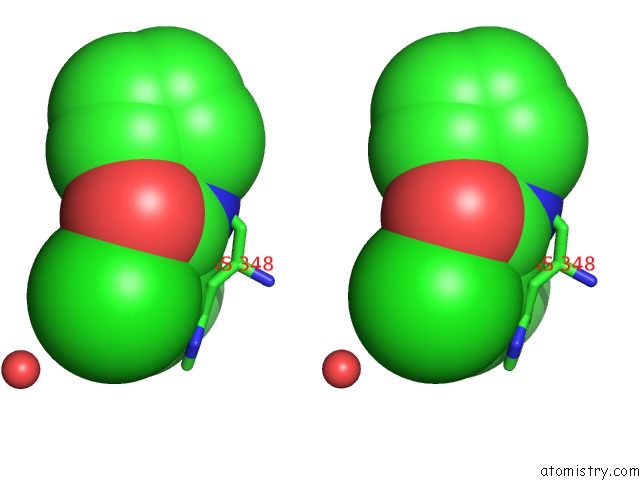
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Pandda Analysis Group Deposition -- Crystal Structure of Human ALAS2A in Complex with Z275151340 within 5.0Å range:
|
Reference:
G.A.Bezerra,
W.Foster,
H.Bailey,
L.Shrestha,
T.Krojer,
R.Talon,
J.Brandao-Neto,
A.Douangamath,
B.B.Nicola,
F.Von Delft,
C.H.Arrowsmith,
A.Edwards,
C.Bountra,
P.E.Brennan,
W.W.Yue.
Pandda Analysis Group Deposition To Be Published.
Page generated: Sat Jul 12 07:45:03 2025
Last articles
Cl in 6B7UCl in 6B5V
Cl in 6B7D
Cl in 6B7L
Cl in 6B7A
Cl in 6B6X
Cl in 6B71
Cl in 6B6U
Cl in 6B6W
Cl in 6B4N