Chlorine »
PDB 5ufo-5up0 »
5uj2 »
Chlorine in PDB 5uj2: Crystal Structure of Hcv NS5B Genotype 2A Jfh-1 Isolate with S15G E86Q E87Q C223H V321I Mutations and DELTA8 Neta Hairpoin Loop Deletion in Complex with Gs-639476 (Diphsohate Version of Gs-9813), MN2+ and Symmetrical Primer Template 5'-Auaaauuu
Protein crystallography data
The structure of Crystal Structure of Hcv NS5B Genotype 2A Jfh-1 Isolate with S15G E86Q E87Q C223H V321I Mutations and DELTA8 Neta Hairpoin Loop Deletion in Complex with Gs-639476 (Diphsohate Version of Gs-9813), MN2+ and Symmetrical Primer Template 5'-Auaaauuu, PDB code: 5uj2
was solved by
T.E.Edwards,
D.Fox Iii,
T.C.Appleby,
E.Murakami,
A.Rey,
M.E.Mcgrath,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 50.00 / 2.90 |
| Space group | P 65 |
| Cell size a, b, c (Å), α, β, γ (°) | 141.760, 141.760, 91.110, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 18.4 / 22.7 |
Other elements in 5uj2:
The structure of Crystal Structure of Hcv NS5B Genotype 2A Jfh-1 Isolate with S15G E86Q E87Q C223H V321I Mutations and DELTA8 Neta Hairpoin Loop Deletion in Complex with Gs-639476 (Diphsohate Version of Gs-9813), MN2+ and Symmetrical Primer Template 5'-Auaaauuu also contains other interesting chemical elements:
| Fluorine | (F) | 1 atom |
| Manganese | (Mn) | 3 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of Hcv NS5B Genotype 2A Jfh-1 Isolate with S15G E86Q E87Q C223H V321I Mutations and DELTA8 Neta Hairpoin Loop Deletion in Complex with Gs-639476 (Diphsohate Version of Gs-9813), MN2+ and Symmetrical Primer Template 5'-Auaaauuu
(pdb code 5uj2). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the Crystal Structure of Hcv NS5B Genotype 2A Jfh-1 Isolate with S15G E86Q E87Q C223H V321I Mutations and DELTA8 Neta Hairpoin Loop Deletion in Complex with Gs-639476 (Diphsohate Version of Gs-9813), MN2+ and Symmetrical Primer Template 5'-Auaaauuu, PDB code: 5uj2:
In total only one binding site of Chlorine was determined in the Crystal Structure of Hcv NS5B Genotype 2A Jfh-1 Isolate with S15G E86Q E87Q C223H V321I Mutations and DELTA8 Neta Hairpoin Loop Deletion in Complex with Gs-639476 (Diphsohate Version of Gs-9813), MN2+ and Symmetrical Primer Template 5'-Auaaauuu, PDB code: 5uj2:
Chlorine binding site 1 out of 1 in 5uj2
Go back to
Chlorine binding site 1 out
of 1 in the Crystal Structure of Hcv NS5B Genotype 2A Jfh-1 Isolate with S15G E86Q E87Q C223H V321I Mutations and DELTA8 Neta Hairpoin Loop Deletion in Complex with Gs-639476 (Diphsohate Version of Gs-9813), MN2+ and Symmetrical Primer Template 5'-Auaaauuu
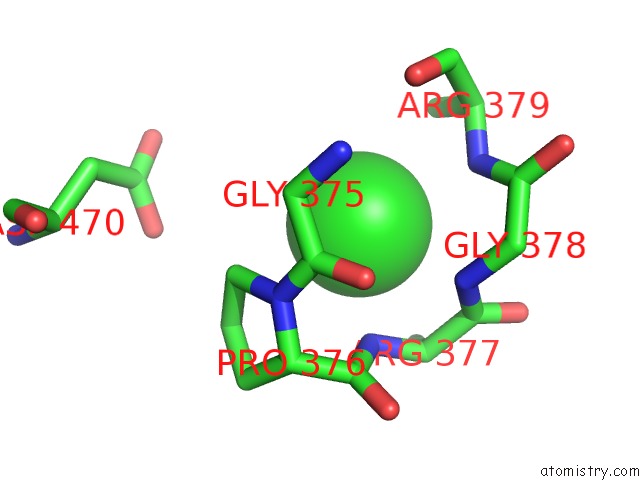
Mono view
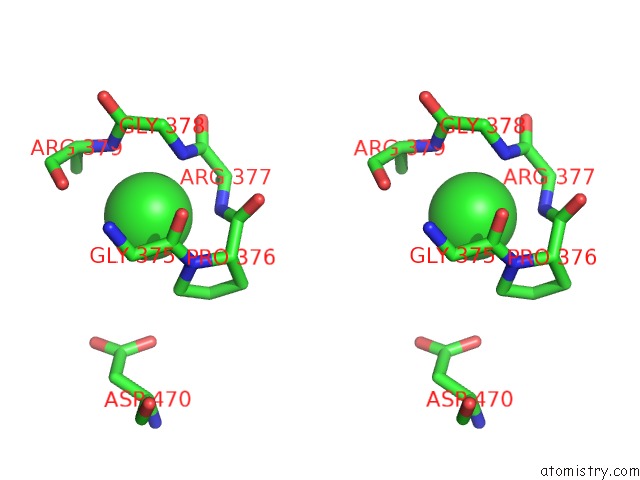
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of Hcv NS5B Genotype 2A Jfh-1 Isolate with S15G E86Q E87Q C223H V321I Mutations and DELTA8 Neta Hairpoin Loop Deletion in Complex with Gs-639476 (Diphsohate Version of Gs-9813), MN2+ and Symmetrical Primer Template 5'-Auaaauuu within 5.0Å range:
|
Reference:
T.A.Kirschberg,
S.Metobo,
M.O.Clarke,
V.Aktoudianakis,
D.Babusis,
O.Barauskas,
G.Birkus,
T.Butler,
D.Byun,
G.Chin,
E.Doerffler,
T.E.Edwards,
M.Fenaux,
R.Lee,
W.Lew,
M.R.Mish,
E.Murakami,
Y.Park,
N.H.Squires,
N.Tirunagari,
T.Wang,
M.Whitcomb,
J.Xu,
H.Yang,
H.Ye,
L.Zhang,
T.C.Appleby,
J.Y.Feng,
A.S.Ray,
A.Cho,
C.U.Kim.
Discovery of A 2'-Fluoro-2'-C-Methyl C-Nucleotide Hcv Polymerase Inhibitor and A Phosphoramidate Prodrug with Favorable Properties. Bioorg. Med. Chem. Lett. V. 27 1840 2017.
ISSN: ESSN 1464-3405
PubMed: 28274633
DOI: 10.1016/J.BMCL.2017.02.037
Page generated: Fri Jul 26 18:07:18 2024
ISSN: ESSN 1464-3405
PubMed: 28274633
DOI: 10.1016/J.BMCL.2017.02.037
Last articles
Zn in 9J0NZn in 9J0O
Zn in 9J0P
Zn in 9FJX
Zn in 9EKB
Zn in 9C0F
Zn in 9CAH
Zn in 9CH0
Zn in 9CH3
Zn in 9CH1