Chlorine »
PDB 5v4h-5vc4 »
5v72 »
Chlorine in PDB 5v72: Crystal Structure of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase SMC04462 (Smghrb) From Sinorhizobium Meliloti in Complex with Citrate
Enzymatic activity of Crystal Structure of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase SMC04462 (Smghrb) From Sinorhizobium Meliloti in Complex with Citrate
All present enzymatic activity of Crystal Structure of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase SMC04462 (Smghrb) From Sinorhizobium Meliloti in Complex with Citrate:
1.1.1.81;
1.1.1.81;
Protein crystallography data
The structure of Crystal Structure of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase SMC04462 (Smghrb) From Sinorhizobium Meliloti in Complex with Citrate, PDB code: 5v72
was solved by
I.G.Shabalin,
K.B.Handing,
O.A.Gasiorowska,
D.R.Cooper,
D.Matelska,
J.Bonanno,
S.C.Almo,
W.Minor,
New York Structural Genomics Researchconsortium (Nysgrc),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 50.00 / 2.10 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 63.188, 157.926, 64.719, 90.00, 110.74, 90.00 |
| R / Rfree (%) | 16.4 / 20.4 |
Other elements in 5v72:
The structure of Crystal Structure of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase SMC04462 (Smghrb) From Sinorhizobium Meliloti in Complex with Citrate also contains other interesting chemical elements:
| Sodium | (Na) | 5 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase SMC04462 (Smghrb) From Sinorhizobium Meliloti in Complex with Citrate
(pdb code 5v72). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 4 binding sites of Chlorine where determined in the Crystal Structure of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase SMC04462 (Smghrb) From Sinorhizobium Meliloti in Complex with Citrate, PDB code: 5v72:
Jump to Chlorine binding site number: 1; 2; 3; 4;
In total 4 binding sites of Chlorine where determined in the Crystal Structure of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase SMC04462 (Smghrb) From Sinorhizobium Meliloti in Complex with Citrate, PDB code: 5v72:
Jump to Chlorine binding site number: 1; 2; 3; 4;
Chlorine binding site 1 out of 4 in 5v72
Go back to
Chlorine binding site 1 out
of 4 in the Crystal Structure of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase SMC04462 (Smghrb) From Sinorhizobium Meliloti in Complex with Citrate
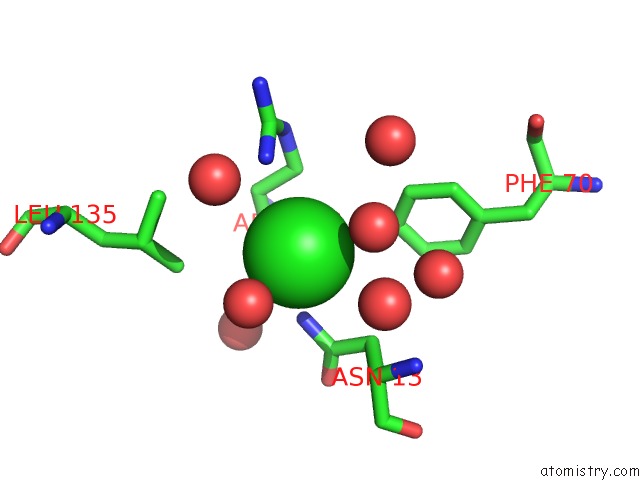
Mono view
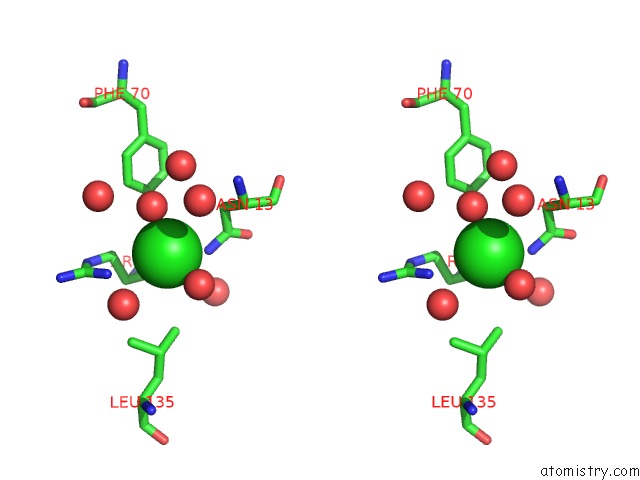
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase SMC04462 (Smghrb) From Sinorhizobium Meliloti in Complex with Citrate within 5.0Å range:
|
Chlorine binding site 2 out of 4 in 5v72
Go back to
Chlorine binding site 2 out
of 4 in the Crystal Structure of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase SMC04462 (Smghrb) From Sinorhizobium Meliloti in Complex with Citrate
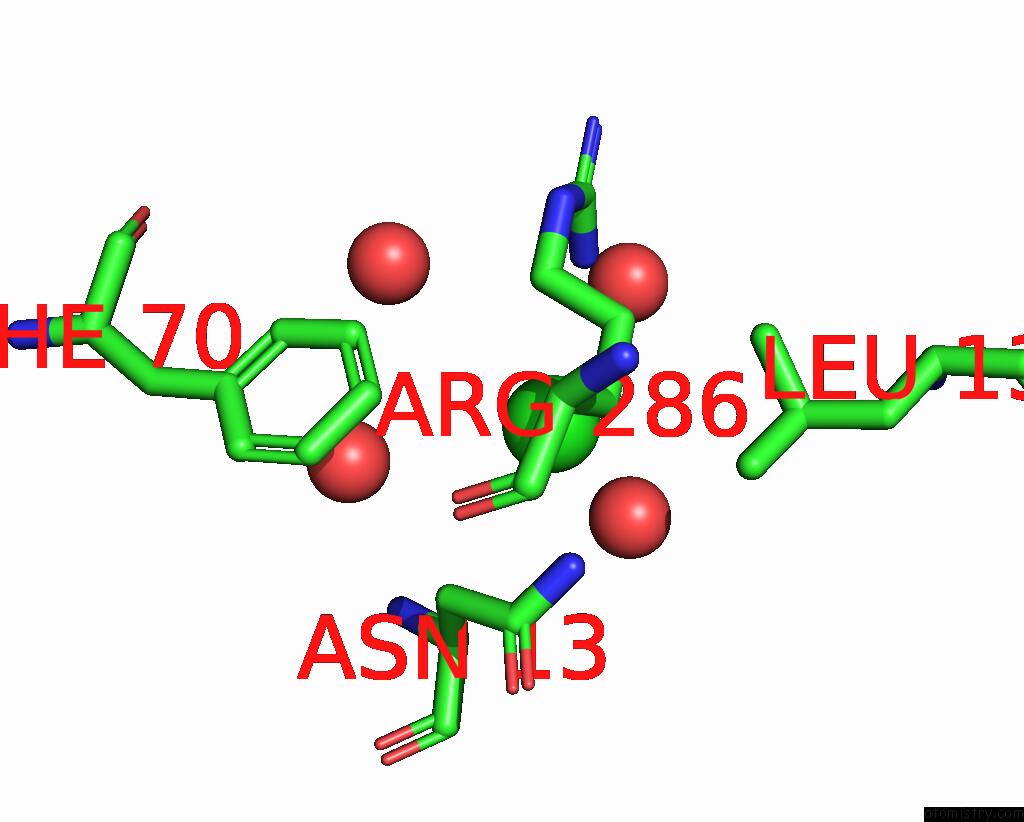
Mono view
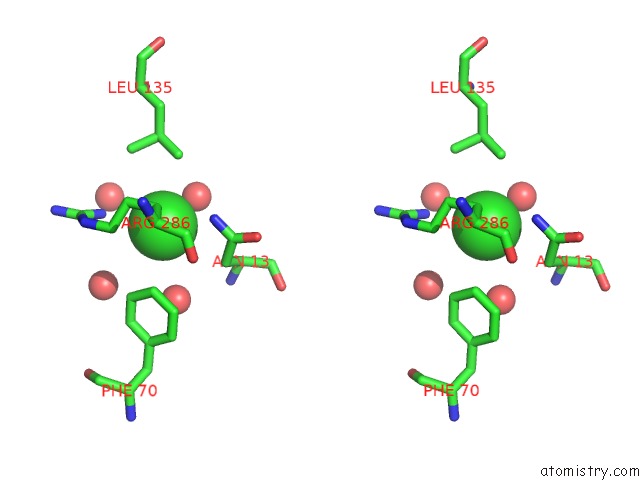
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase SMC04462 (Smghrb) From Sinorhizobium Meliloti in Complex with Citrate within 5.0Å range:
|
Chlorine binding site 3 out of 4 in 5v72
Go back to
Chlorine binding site 3 out
of 4 in the Crystal Structure of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase SMC04462 (Smghrb) From Sinorhizobium Meliloti in Complex with Citrate
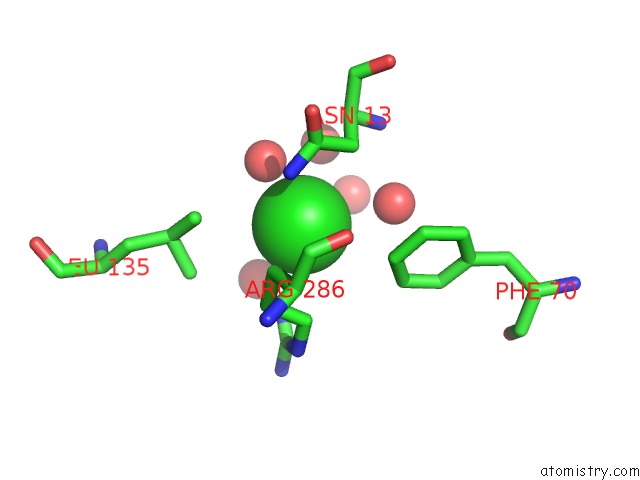
Mono view
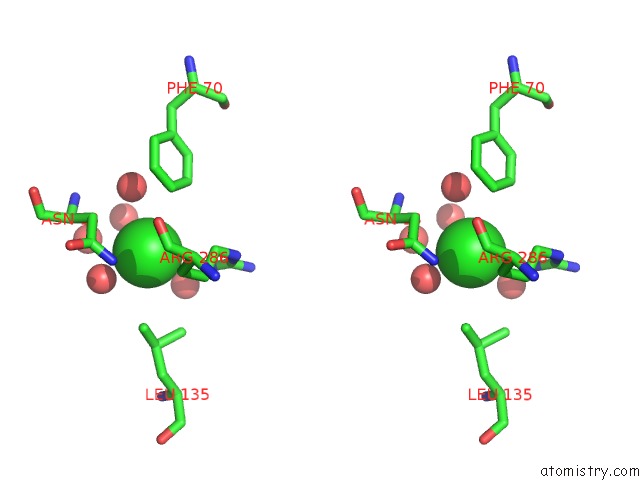
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Crystal Structure of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase SMC04462 (Smghrb) From Sinorhizobium Meliloti in Complex with Citrate within 5.0Å range:
|
Chlorine binding site 4 out of 4 in 5v72
Go back to
Chlorine binding site 4 out
of 4 in the Crystal Structure of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase SMC04462 (Smghrb) From Sinorhizobium Meliloti in Complex with Citrate
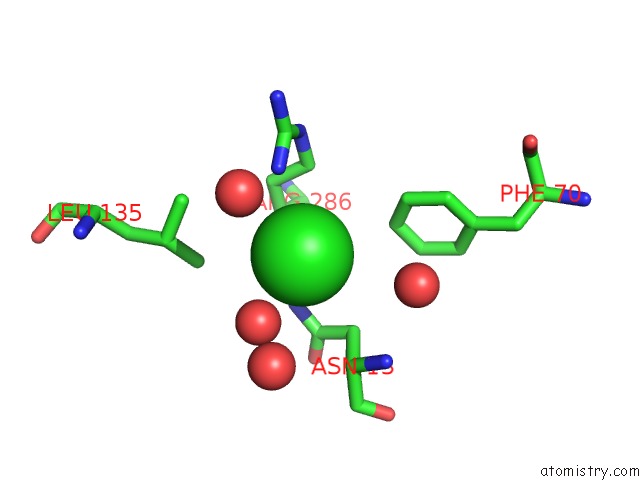
Mono view
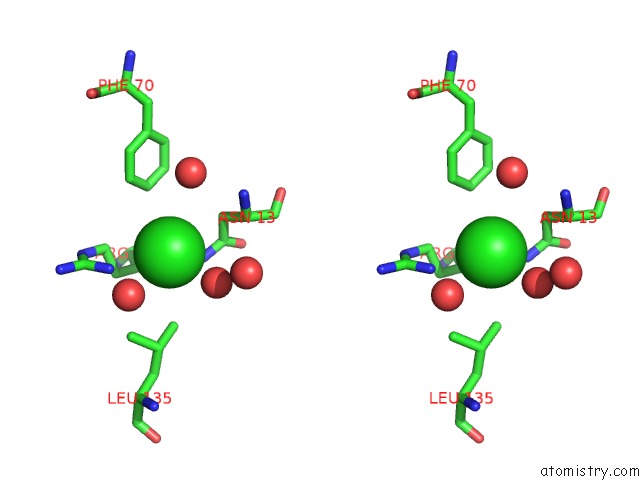
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Crystal Structure of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase SMC04462 (Smghrb) From Sinorhizobium Meliloti in Complex with Citrate within 5.0Å range:
|
Reference:
J.Kutner,
I.G.Shabalin,
D.Matelska,
K.B.Handing,
O.Gasiorowska,
P.Sroka,
M.W.Gorna,
K.Ginalski,
K.Wozniak,
W.Minor.
Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division Into Two Distinct Subfamilies. Biochemistry V. 57 963 2018.
ISSN: ISSN 1520-4995
PubMed: 29309127
DOI: 10.1021/ACS.BIOCHEM.7B01137
Page generated: Sat Jul 12 09:45:14 2025
ISSN: ISSN 1520-4995
PubMed: 29309127
DOI: 10.1021/ACS.BIOCHEM.7B01137
Last articles
F in 7RJ4F in 7RJ2
F in 7RHN
F in 7RME
F in 7RKW
F in 7RKT
F in 7RKR
F in 7RJE
F in 7RJ8
F in 7RH7