Chlorine »
PDB 5y7x-5yka »
5yiy »
Chlorine in PDB 5yiy: Crystal Structure of D175A Mutant of RV3272 From Mycobacterium Tuberculosis
Protein crystallography data
The structure of Crystal Structure of D175A Mutant of RV3272 From Mycobacterium Tuberculosis, PDB code: 5yiy
was solved by
S.S.Karade,
J.V.Pratap,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 49.12 / 2.50 |
| Space group | P 43 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 78.678, 78.678, 251.475, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20.2 / 23.5 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of D175A Mutant of RV3272 From Mycobacterium Tuberculosis
(pdb code 5yiy). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the Crystal Structure of D175A Mutant of RV3272 From Mycobacterium Tuberculosis, PDB code: 5yiy:
In total only one binding site of Chlorine was determined in the Crystal Structure of D175A Mutant of RV3272 From Mycobacterium Tuberculosis, PDB code: 5yiy:
Chlorine binding site 1 out of 1 in 5yiy
Go back to
Chlorine binding site 1 out
of 1 in the Crystal Structure of D175A Mutant of RV3272 From Mycobacterium Tuberculosis
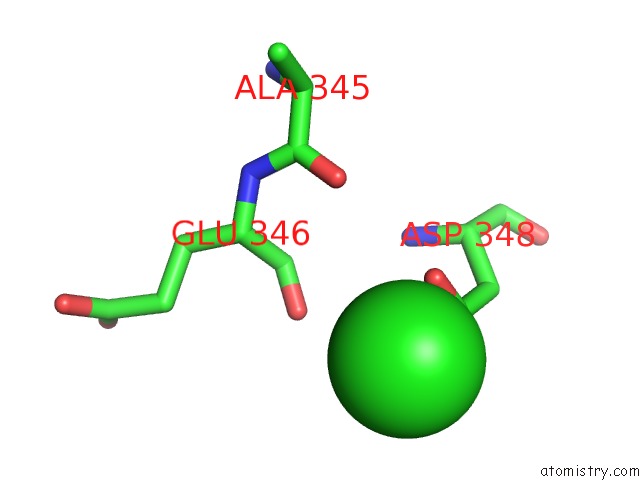
Mono view
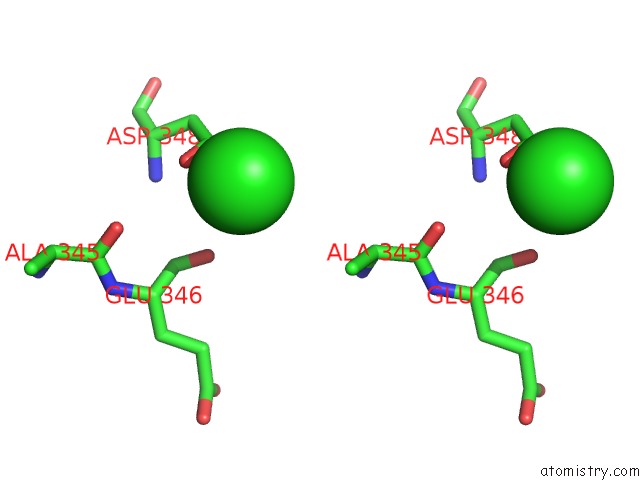
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of D175A Mutant of RV3272 From Mycobacterium Tuberculosis within 5.0Å range:
|
Reference:
S.S.Karade,
S.Pandey,
A.Ansari,
S.Das,
S.Tripathi,
A.Arora,
S.Chopra,
J.V.Pratap,
A.Dasgupta.
RV3272 Encodes A Novel Family III Coa Transferase That Alters the Cell Wall Lipid Profile and Protects Mycobacteria From Acidic and Oxidative Stress. Biochim Biophys Acta V.1867 317 2019PROTEINS Proteom.
ISSN: ISSN 1878-1454
PubMed: 30342240
DOI: 10.1016/J.BBAPAP.2018.10.011
Page generated: Fri Jul 26 21:26:06 2024
ISSN: ISSN 1878-1454
PubMed: 30342240
DOI: 10.1016/J.BBAPAP.2018.10.011
Last articles
Zn in 9J0NZn in 9J0O
Zn in 9J0P
Zn in 9FJX
Zn in 9EKB
Zn in 9C0F
Zn in 9CAH
Zn in 9CH0
Zn in 9CH3
Zn in 9CH1