Chlorine »
PDB 6b4n-6bb2 »
6baa »
Chlorine in PDB 6baa: Cryo-Em Structure of the Pancreatic Beta-Cell Katp Channel Bound to Atp and Glibenclamide
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Cryo-Em Structure of the Pancreatic Beta-Cell Katp Channel Bound to Atp and Glibenclamide
(pdb code 6baa). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 4 binding sites of Chlorine where determined in the Cryo-Em Structure of the Pancreatic Beta-Cell Katp Channel Bound to Atp and Glibenclamide, PDB code: 6baa:
Jump to Chlorine binding site number: 1; 2; 3; 4;
In total 4 binding sites of Chlorine where determined in the Cryo-Em Structure of the Pancreatic Beta-Cell Katp Channel Bound to Atp and Glibenclamide, PDB code: 6baa:
Jump to Chlorine binding site number: 1; 2; 3; 4;
Chlorine binding site 1 out of 4 in 6baa
Go back to
Chlorine binding site 1 out
of 4 in the Cryo-Em Structure of the Pancreatic Beta-Cell Katp Channel Bound to Atp and Glibenclamide

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Cryo-Em Structure of the Pancreatic Beta-Cell Katp Channel Bound to Atp and Glibenclamide within 5.0Å range:
|
Chlorine binding site 2 out of 4 in 6baa
Go back to
Chlorine binding site 2 out
of 4 in the Cryo-Em Structure of the Pancreatic Beta-Cell Katp Channel Bound to Atp and Glibenclamide

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Cryo-Em Structure of the Pancreatic Beta-Cell Katp Channel Bound to Atp and Glibenclamide within 5.0Å range:
|
Chlorine binding site 3 out of 4 in 6baa
Go back to
Chlorine binding site 3 out
of 4 in the Cryo-Em Structure of the Pancreatic Beta-Cell Katp Channel Bound to Atp and Glibenclamide

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Cryo-Em Structure of the Pancreatic Beta-Cell Katp Channel Bound to Atp and Glibenclamide within 5.0Å range:
|
Chlorine binding site 4 out of 4 in 6baa
Go back to
Chlorine binding site 4 out
of 4 in the Cryo-Em Structure of the Pancreatic Beta-Cell Katp Channel Bound to Atp and Glibenclamide
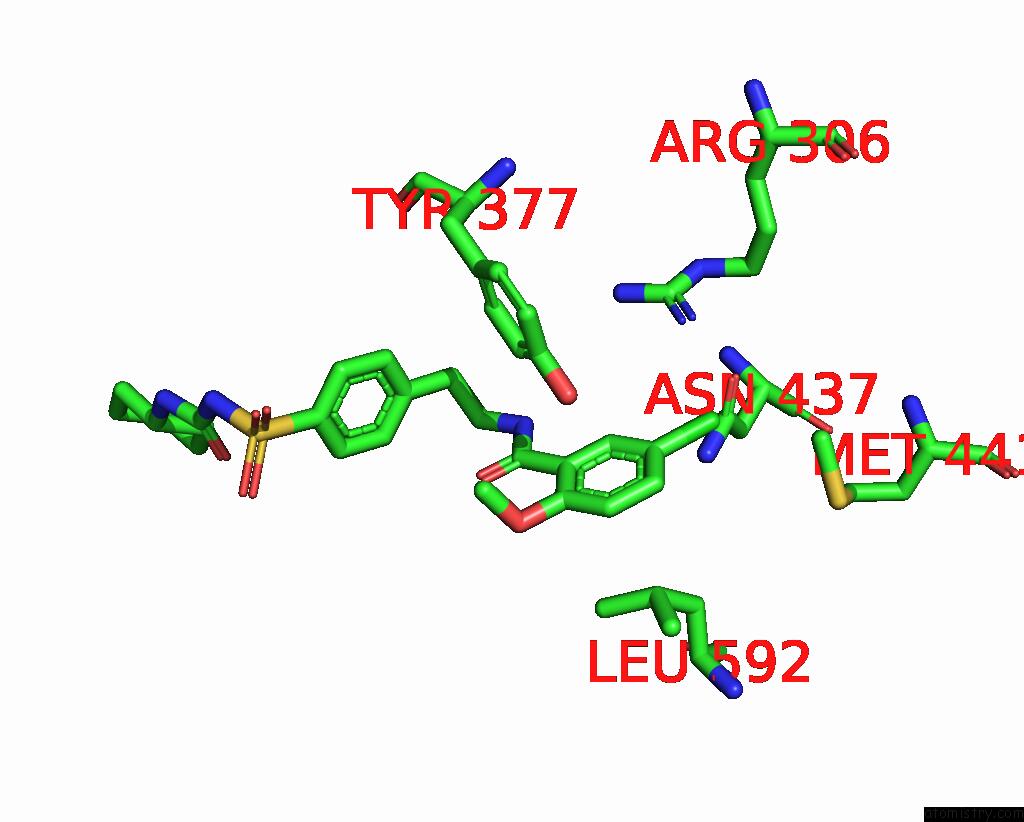
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Cryo-Em Structure of the Pancreatic Beta-Cell Katp Channel Bound to Atp and Glibenclamide within 5.0Å range:
|
Reference:
G.M.Martin,
B.Kandasamy,
F.Dimaio,
C.Yoshioka,
S.L.Shyng.
Anti-Diabetic Drug Binding Site in A Mammalian Katpchannel Revealed By Cryo-Em. Elife V. 6 2017.
ISSN: ESSN 2050-084X
PubMed: 29035201
DOI: 10.7554/ELIFE.31054
Page generated: Fri Jul 26 22:39:47 2024
ISSN: ESSN 2050-084X
PubMed: 29035201
DOI: 10.7554/ELIFE.31054
Last articles
Cl in 2X0RCl in 2X1C
Cl in 2X1E
Cl in 2X10
Cl in 2X0A
Cl in 2WZZ
Cl in 2WZN
Cl in 2WZX
Cl in 2WZD
Cl in 2WZQ