Chlorine »
PDB 6cdr-6cod »
6clz »
Chlorine in PDB 6clz: MT1-Mmp Hpx Domain with Blade 4 Loop Bound to Nanodiscs
Enzymatic activity of MT1-Mmp Hpx Domain with Blade 4 Loop Bound to Nanodiscs
All present enzymatic activity of MT1-Mmp Hpx Domain with Blade 4 Loop Bound to Nanodiscs:
3.4.24.80;
3.4.24.80;
Other elements in 6clz:
The structure of MT1-Mmp Hpx Domain with Blade 4 Loop Bound to Nanodiscs also contains other interesting chemical elements:
| Sodium | (Na) | 15 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the MT1-Mmp Hpx Domain with Blade 4 Loop Bound to Nanodiscs
(pdb code 6clz). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the MT1-Mmp Hpx Domain with Blade 4 Loop Bound to Nanodiscs, PDB code: 6clz:
In total only one binding site of Chlorine was determined in the MT1-Mmp Hpx Domain with Blade 4 Loop Bound to Nanodiscs, PDB code: 6clz:
Chlorine binding site 1 out of 1 in 6clz
Go back to
Chlorine binding site 1 out
of 1 in the MT1-Mmp Hpx Domain with Blade 4 Loop Bound to Nanodiscs
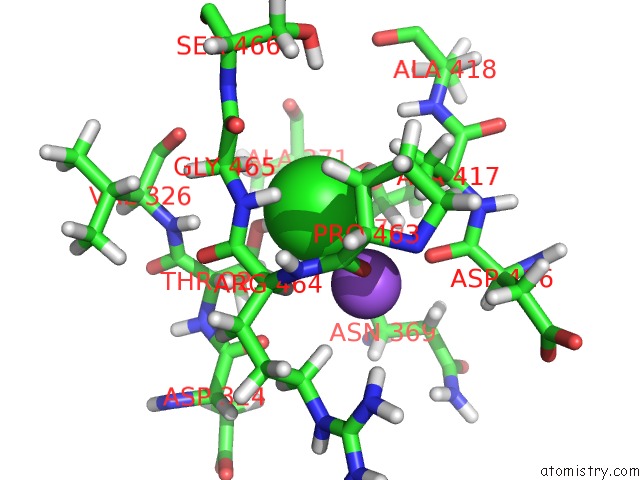
Mono view
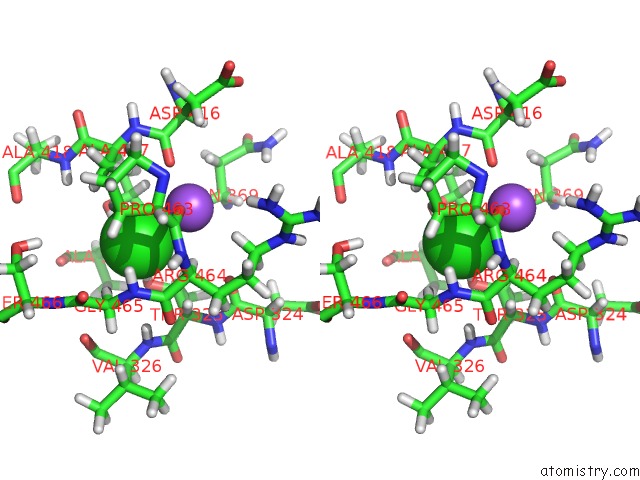
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of MT1-Mmp Hpx Domain with Blade 4 Loop Bound to Nanodiscs within 5.0Å range:
|
Reference:
T.C.Marcink,
J.A.Simoncic,
B.An,
A.M.Knapinska,
Y.G.Fulcher,
N.Akkaladevi,
G.B.Fields,
S.R.Van Doren.
MT1-Mmp Binds Membranes By Opposite Tips of Its Beta Propeller to Position It For Pericellular Proteolysis. Structure V. 27 281 2019.
ISSN: ISSN 1878-4186
PubMed: 30471921
DOI: 10.1016/J.STR.2018.10.008
Page generated: Fri Jul 26 23:35:53 2024
ISSN: ISSN 1878-4186
PubMed: 30471921
DOI: 10.1016/J.STR.2018.10.008
Last articles
Zn in 9J0NZn in 9J0O
Zn in 9J0P
Zn in 9FJX
Zn in 9EKB
Zn in 9C0F
Zn in 9CAH
Zn in 9CH0
Zn in 9CH3
Zn in 9CH1