Chlorine »
PDB 6dow-6dy1 »
6dy0 »
Chlorine in PDB 6dy0: Rabbit N-Acylethanolamine-Hydrolyzing Acid Amidase (Naaa) Covalently Bound to Beta-Lactam Inhibitor ARN726, in Presence of Triton X-100
Enzymatic activity of Rabbit N-Acylethanolamine-Hydrolyzing Acid Amidase (Naaa) Covalently Bound to Beta-Lactam Inhibitor ARN726, in Presence of Triton X-100
All present enzymatic activity of Rabbit N-Acylethanolamine-Hydrolyzing Acid Amidase (Naaa) Covalently Bound to Beta-Lactam Inhibitor ARN726, in Presence of Triton X-100:
3.5.1.60;
3.5.1.60;
Protein crystallography data
The structure of Rabbit N-Acylethanolamine-Hydrolyzing Acid Amidase (Naaa) Covalently Bound to Beta-Lactam Inhibitor ARN726, in Presence of Triton X-100, PDB code: 6dy0
was solved by
A.Gorelik,
A.Gebai,
K.Illes,
D.Piomelli,
B.Nagar,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 49.92 / 3.01 |
| Space group | P 43 3 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 157.871, 157.871, 157.871, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 19.4 / 25 |
Chlorine Binding Sites:
Pages:
>>> Page 1 <<< Page 2, Binding sites: 11 - 12;Binding sites:
The binding sites of Chlorine atom in the Rabbit N-Acylethanolamine-Hydrolyzing Acid Amidase (Naaa) Covalently Bound to Beta-Lactam Inhibitor ARN726, in Presence of Triton X-100 (pdb code 6dy0). This binding sites where shown within 5.0 Angstroms radius around Chlorine atom.In total 12 binding sites of Chlorine where determined in the Rabbit N-Acylethanolamine-Hydrolyzing Acid Amidase (Naaa) Covalently Bound to Beta-Lactam Inhibitor ARN726, in Presence of Triton X-100, PDB code: 6dy0:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9; 10;
Chlorine binding site 1 out of 12 in 6dy0
Go back to
Chlorine binding site 1 out
of 12 in the Rabbit N-Acylethanolamine-Hydrolyzing Acid Amidase (Naaa) Covalently Bound to Beta-Lactam Inhibitor ARN726, in Presence of Triton X-100

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Rabbit N-Acylethanolamine-Hydrolyzing Acid Amidase (Naaa) Covalently Bound to Beta-Lactam Inhibitor ARN726, in Presence of Triton X-100 within 5.0Å range:
|
Chlorine binding site 2 out of 12 in 6dy0
Go back to
Chlorine binding site 2 out
of 12 in the Rabbit N-Acylethanolamine-Hydrolyzing Acid Amidase (Naaa) Covalently Bound to Beta-Lactam Inhibitor ARN726, in Presence of Triton X-100

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Rabbit N-Acylethanolamine-Hydrolyzing Acid Amidase (Naaa) Covalently Bound to Beta-Lactam Inhibitor ARN726, in Presence of Triton X-100 within 5.0Å range:
|
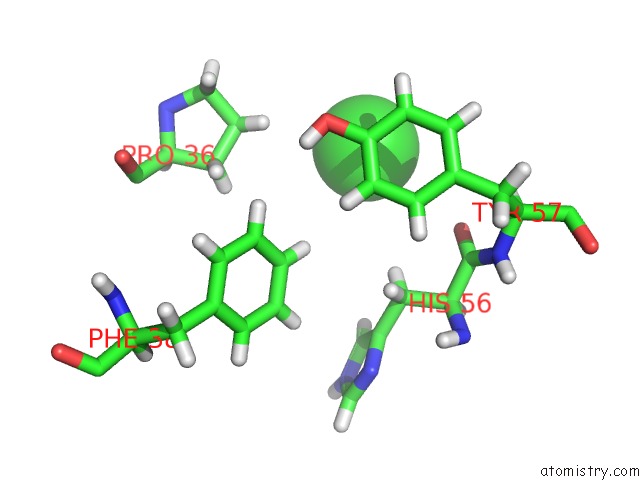
Chlorine binding site 3 out of 12 in 6dy0
Go back to
Chlorine binding site 3 out
of 12 in the Rabbit N-Acylethanolamine-Hydrolyzing Acid Amidase (Naaa) Covalently Bound to Beta-Lactam Inhibitor ARN726, in Presence of Triton X-100
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Rabbit N-Acylethanolamine-Hydrolyzing Acid Amidase (Naaa) Covalently Bound to Beta-Lactam Inhibitor ARN726, in Presence of Triton X-100 within 5.0Å range:
|
Chlorine binding site 4 out of 12 in 6dy0
Go back to
Chlorine binding site 4 out
of 12 in the Rabbit N-Acylethanolamine-Hydrolyzing Acid Amidase (Naaa) Covalently Bound to Beta-Lactam Inhibitor ARN726, in Presence of Triton X-100

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Rabbit N-Acylethanolamine-Hydrolyzing Acid Amidase (Naaa) Covalently Bound to Beta-Lactam Inhibitor ARN726, in Presence of Triton X-100 within 5.0Å range:
|
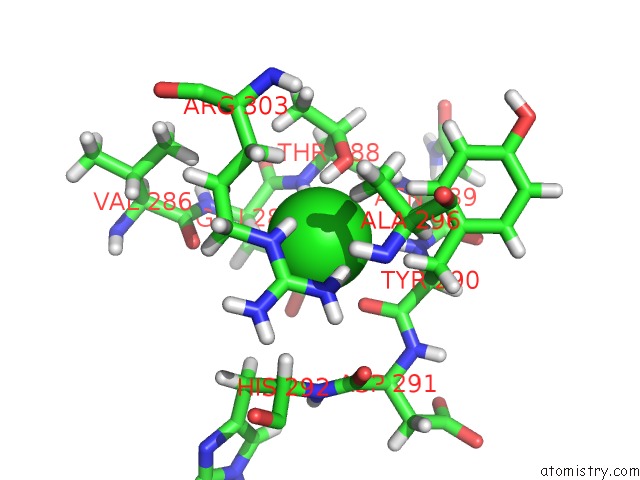
Chlorine binding site 5 out of 12 in 6dy0
Go back to
Chlorine binding site 5 out
of 12 in the Rabbit N-Acylethanolamine-Hydrolyzing Acid Amidase (Naaa) Covalently Bound to Beta-Lactam Inhibitor ARN726, in Presence of Triton X-100
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Rabbit N-Acylethanolamine-Hydrolyzing Acid Amidase (Naaa) Covalently Bound to Beta-Lactam Inhibitor ARN726, in Presence of Triton X-100 within 5.0Å range:
|
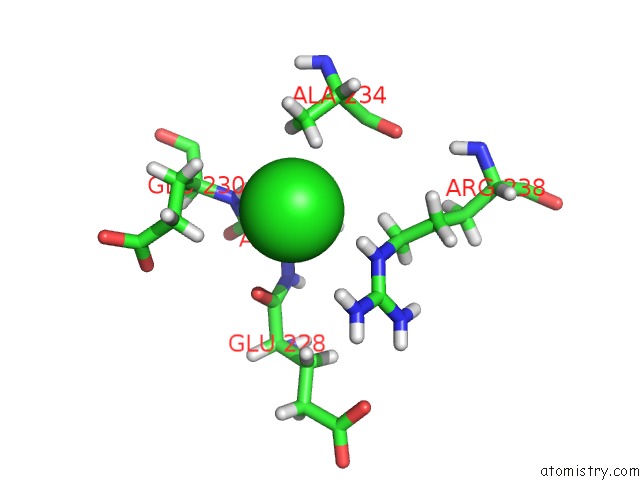
Chlorine binding site 6 out of 12 in 6dy0
Go back to
Chlorine binding site 6 out
of 12 in the Rabbit N-Acylethanolamine-Hydrolyzing Acid Amidase (Naaa) Covalently Bound to Beta-Lactam Inhibitor ARN726, in Presence of Triton X-100
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 6 of Rabbit N-Acylethanolamine-Hydrolyzing Acid Amidase (Naaa) Covalently Bound to Beta-Lactam Inhibitor ARN726, in Presence of Triton X-100 within 5.0Å range:
|
Chlorine binding site 7 out of 12 in 6dy0
Go back to
Chlorine binding site 7 out
of 12 in the Rabbit N-Acylethanolamine-Hydrolyzing Acid Amidase (Naaa) Covalently Bound to Beta-Lactam Inhibitor ARN726, in Presence of Triton X-100

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 7 of Rabbit N-Acylethanolamine-Hydrolyzing Acid Amidase (Naaa) Covalently Bound to Beta-Lactam Inhibitor ARN726, in Presence of Triton X-100 within 5.0Å range:
|
Chlorine binding site 8 out of 12 in 6dy0
Go back to
Chlorine binding site 8 out
of 12 in the Rabbit N-Acylethanolamine-Hydrolyzing Acid Amidase (Naaa) Covalently Bound to Beta-Lactam Inhibitor ARN726, in Presence of Triton X-100

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 8 of Rabbit N-Acylethanolamine-Hydrolyzing Acid Amidase (Naaa) Covalently Bound to Beta-Lactam Inhibitor ARN726, in Presence of Triton X-100 within 5.0Å range:
|
Chlorine binding site 9 out of 12 in 6dy0
Go back to
Chlorine binding site 9 out
of 12 in the Rabbit N-Acylethanolamine-Hydrolyzing Acid Amidase (Naaa) Covalently Bound to Beta-Lactam Inhibitor ARN726, in Presence of Triton X-100

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 9 of Rabbit N-Acylethanolamine-Hydrolyzing Acid Amidase (Naaa) Covalently Bound to Beta-Lactam Inhibitor ARN726, in Presence of Triton X-100 within 5.0Å range:
|
Chlorine binding site 10 out of 12 in 6dy0
Go back to
Chlorine binding site 10 out
of 12 in the Rabbit N-Acylethanolamine-Hydrolyzing Acid Amidase (Naaa) Covalently Bound to Beta-Lactam Inhibitor ARN726, in Presence of Triton X-100

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 10 of Rabbit N-Acylethanolamine-Hydrolyzing Acid Amidase (Naaa) Covalently Bound to Beta-Lactam Inhibitor ARN726, in Presence of Triton X-100 within 5.0Å range:
|
Reference:
A.Gorelik,
A.Gebai,
K.Illes,
D.Piomelli,
B.Nagar.
Molecular Mechanism of Activation of the Immunoregulatory Amidase Naaa. Proc. Natl. Acad. Sci. V. 115 10032 2018U.S.A..
ISSN: ESSN 1091-6490
PubMed: 30301806
DOI: 10.1073/PNAS.1811759115
Page generated: Sat Jul 27 21:52:35 2024
ISSN: ESSN 1091-6490
PubMed: 30301806
DOI: 10.1073/PNAS.1811759115
Last articles
Ca in 2TN4Ca in 2USN
Ca in 2UUY
Ca in 2UUK
Ca in 2UUJ
Ca in 2UUF
Ca in 2UU8
Ca in 2TRM
Ca in 2TMN
Ca in 2TLI