Chlorine »
PDB 6fbv-6fkv »
6fcj »
Chlorine in PDB 6fcj: Estimation of Protein-Ligand Unbinding Kinetics Using Non-Equilibrium Targeted Molecular Dynamics Simulations
Protein crystallography data
The structure of Estimation of Protein-Ligand Unbinding Kinetics Using Non-Equilibrium Targeted Molecular Dynamics Simulations, PDB code: 6fcj
was solved by
D.Musil,
M.Lehmann,
H.-M.Eggenweiler,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 33.15 / 2.49 |
| Space group | I 2 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 66.015, 89.814, 98.470, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 18.8 / 23.4 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Estimation of Protein-Ligand Unbinding Kinetics Using Non-Equilibrium Targeted Molecular Dynamics Simulations
(pdb code 6fcj). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Estimation of Protein-Ligand Unbinding Kinetics Using Non-Equilibrium Targeted Molecular Dynamics Simulations, PDB code: 6fcj:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Estimation of Protein-Ligand Unbinding Kinetics Using Non-Equilibrium Targeted Molecular Dynamics Simulations, PDB code: 6fcj:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 6fcj
Go back to
Chlorine binding site 1 out
of 2 in the Estimation of Protein-Ligand Unbinding Kinetics Using Non-Equilibrium Targeted Molecular Dynamics Simulations
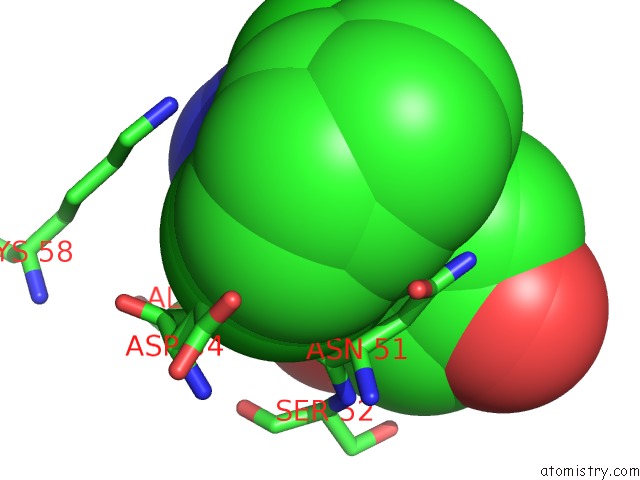
Mono view
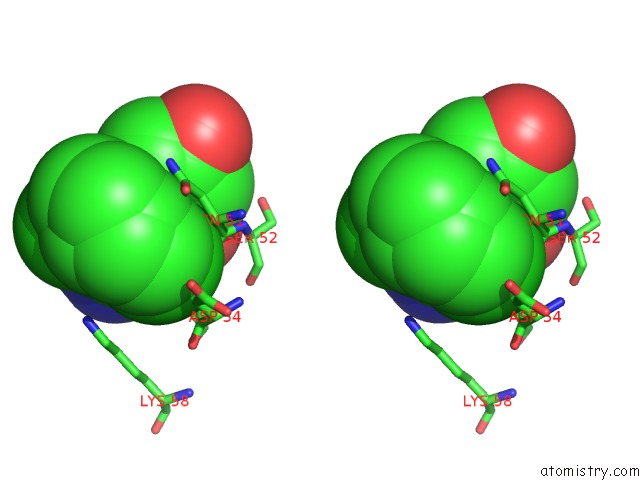
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Estimation of Protein-Ligand Unbinding Kinetics Using Non-Equilibrium Targeted Molecular Dynamics Simulations within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 6fcj
Go back to
Chlorine binding site 2 out
of 2 in the Estimation of Protein-Ligand Unbinding Kinetics Using Non-Equilibrium Targeted Molecular Dynamics Simulations
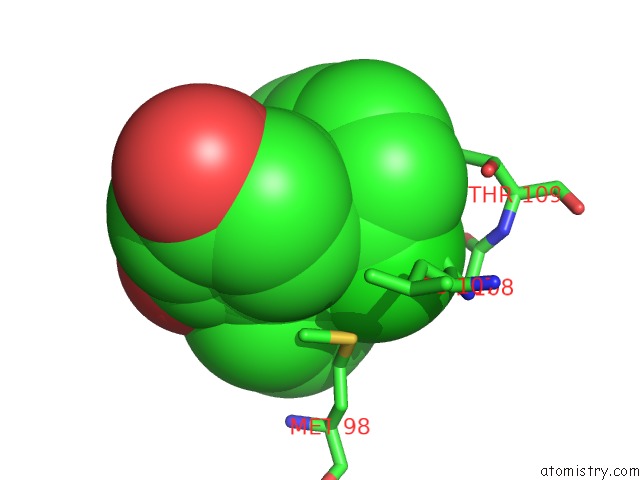
Mono view
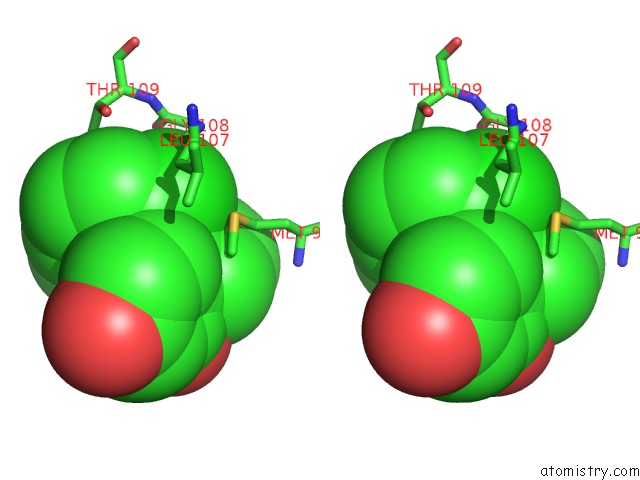
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Estimation of Protein-Ligand Unbinding Kinetics Using Non-Equilibrium Targeted Molecular Dynamics Simulations within 5.0Å range:
|
Reference:
S.Wolf,
M.Amaral,
M.Lowinski,
F.Vallee,
D.Musil,
J.Guldenhaupt,
M.K.Dreyer,
J.Bomke,
M.Frech,
J.Schlitter,
K.Gerwert.
Estimation of Protein-Ligand Unbinding Kinetics Using Non-Equilibrium Targeted Molecular Dynamics Simulations Arxiv 2019.
ISSN: ISSN 2331-8422
Page generated: Sat Jul 12 13:59:05 2025
ISSN: ISSN 2331-8422
Last articles
F in 4I5XF in 4I5H
F in 4I54
F in 4I53
F in 4I24
F in 4I23
F in 4I0I
F in 4I22
F in 4I0H
F in 4I0J