Chlorine »
PDB 6fl5-6frq »
6fo0 »
Chlorine in PDB 6fo0: Cryoem Structure of Bovine Cytochrome BC1 in Complex with the Anti- Malarial Compound GSK932121
Enzymatic activity of Cryoem Structure of Bovine Cytochrome BC1 in Complex with the Anti- Malarial Compound GSK932121
All present enzymatic activity of Cryoem Structure of Bovine Cytochrome BC1 in Complex with the Anti- Malarial Compound GSK932121:
1.10.2.2;
1.10.2.2;
Other elements in 6fo0:
The structure of Cryoem Structure of Bovine Cytochrome BC1 in Complex with the Anti- Malarial Compound GSK932121 also contains other interesting chemical elements:
| Fluorine | (F) | 6 atoms |
| Iron | (Fe) | 6 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Cryoem Structure of Bovine Cytochrome BC1 in Complex with the Anti- Malarial Compound GSK932121
(pdb code 6fo0). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Cryoem Structure of Bovine Cytochrome BC1 in Complex with the Anti- Malarial Compound GSK932121, PDB code: 6fo0:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Cryoem Structure of Bovine Cytochrome BC1 in Complex with the Anti- Malarial Compound GSK932121, PDB code: 6fo0:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 6fo0
Go back to
Chlorine binding site 1 out
of 2 in the Cryoem Structure of Bovine Cytochrome BC1 in Complex with the Anti- Malarial Compound GSK932121
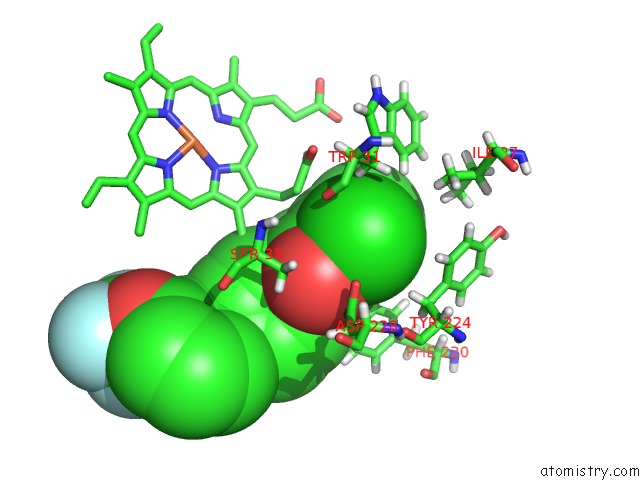
Mono view
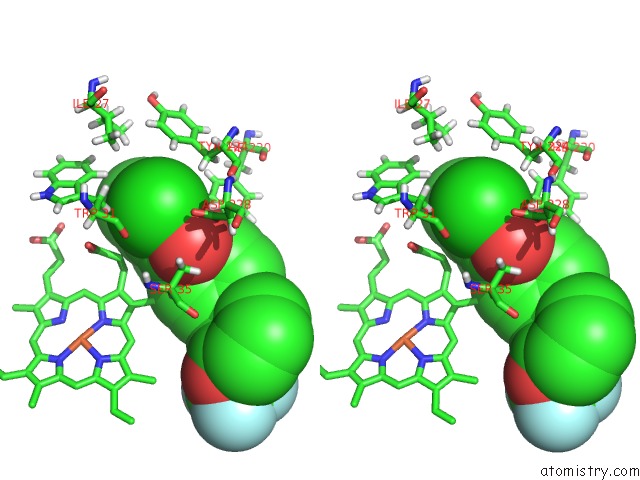
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Cryoem Structure of Bovine Cytochrome BC1 in Complex with the Anti- Malarial Compound GSK932121 within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 6fo0
Go back to
Chlorine binding site 2 out
of 2 in the Cryoem Structure of Bovine Cytochrome BC1 in Complex with the Anti- Malarial Compound GSK932121
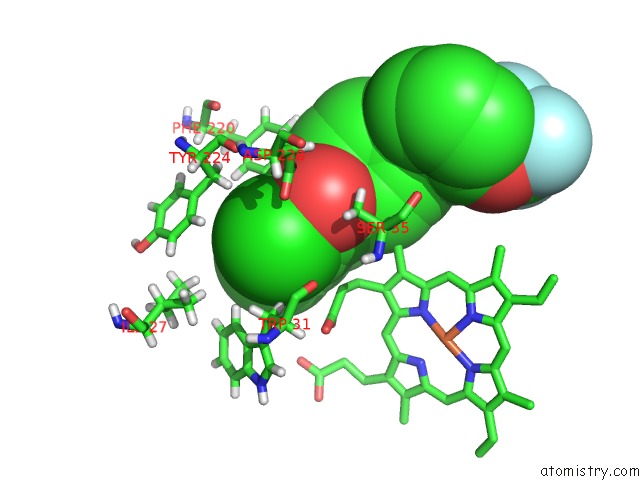
Mono view
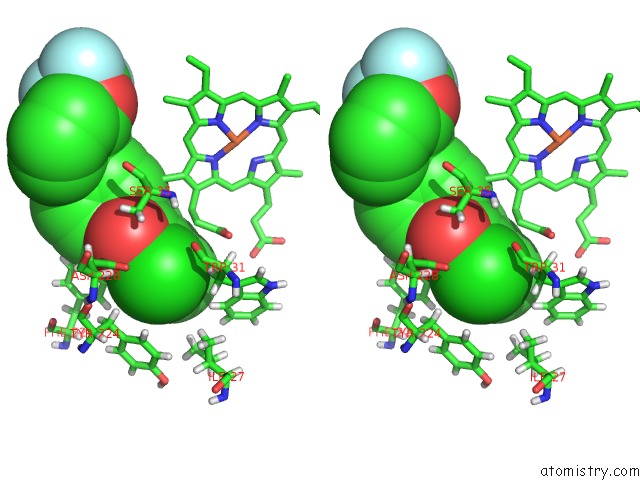
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Cryoem Structure of Bovine Cytochrome BC1 in Complex with the Anti- Malarial Compound GSK932121 within 5.0Å range:
|
Reference:
K.Amporndanai,
R.M.Johnson,
P.M.O'neill,
C.W.G.Fishwick,
A.H.Jamson,
S.Rawson,
S.P.Muench,
S.S.Hasnain,
S.V.Antonyuk.
X-Ray and Cryo-Em Structures of Inhibitor-Bound CYTOCHROMEBC1COMPLEXES For Structure-Based Drug Discovery. Iucrj V. 5 200 2018.
ISSN: ESSN 2052-2525
PubMed: 29765610
DOI: 10.1107/S2052252518001616
Page generated: Sat Jul 27 23:23:45 2024
ISSN: ESSN 2052-2525
PubMed: 29765610
DOI: 10.1107/S2052252518001616
Last articles
Cl in 2W79Cl in 2W5P
Cl in 2W6Q
Cl in 2W6Z
Cl in 2W6O
Cl in 2W6N
Cl in 2W4L
Cl in 2W6M
Cl in 2W5B
Cl in 2W4M