Chlorine »
PDB 6ggo-6gos »
6gim »
Chlorine in PDB 6gim: Structure of the Dna Duplex D(Aaattt)2 with [N-(3-Chloro-4-((4,5- Dihydro-1H-Imidazol-2-Yl)Amino)Phenyl)-4-((4,5-Dihydro-1H-Imidazol-2- Yl)Amino)Benzamide] - (Drug JNI18)
Protein crystallography data
The structure of Structure of the Dna Duplex D(Aaattt)2 with [N-(3-Chloro-4-((4,5- Dihydro-1H-Imidazol-2-Yl)Amino)Phenyl)-4-((4,5-Dihydro-1H-Imidazol-2- Yl)Amino)Benzamide] - (Drug JNI18), PDB code: 6gim
was solved by
C.R.Millan,
C.Dardonvile,
H.P.De Koning,
N.Saperas,
J.L.Campos,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 36.04 / 1.43 |
| Space group | I 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 22.630, 40.470, 72.255, 90.00, 93.91, 90.00 |
| R / Rfree (%) | 14.3 / 19.3 |
Other elements in 6gim:
The structure of Structure of the Dna Duplex D(Aaattt)2 with [N-(3-Chloro-4-((4,5- Dihydro-1H-Imidazol-2-Yl)Amino)Phenyl)-4-((4,5-Dihydro-1H-Imidazol-2- Yl)Amino)Benzamide] - (Drug JNI18) also contains other interesting chemical elements:
| Magnesium | (Mg) | 3 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Structure of the Dna Duplex D(Aaattt)2 with [N-(3-Chloro-4-((4,5- Dihydro-1H-Imidazol-2-Yl)Amino)Phenyl)-4-((4,5-Dihydro-1H-Imidazol-2- Yl)Amino)Benzamide] - (Drug JNI18)
(pdb code 6gim). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 3 binding sites of Chlorine where determined in the Structure of the Dna Duplex D(Aaattt)2 with [N-(3-Chloro-4-((4,5- Dihydro-1H-Imidazol-2-Yl)Amino)Phenyl)-4-((4,5-Dihydro-1H-Imidazol-2- Yl)Amino)Benzamide] - (Drug JNI18), PDB code: 6gim:
Jump to Chlorine binding site number: 1; 2; 3;
In total 3 binding sites of Chlorine where determined in the Structure of the Dna Duplex D(Aaattt)2 with [N-(3-Chloro-4-((4,5- Dihydro-1H-Imidazol-2-Yl)Amino)Phenyl)-4-((4,5-Dihydro-1H-Imidazol-2- Yl)Amino)Benzamide] - (Drug JNI18), PDB code: 6gim:
Jump to Chlorine binding site number: 1; 2; 3;
Chlorine binding site 1 out of 3 in 6gim
Go back to
Chlorine binding site 1 out
of 3 in the Structure of the Dna Duplex D(Aaattt)2 with [N-(3-Chloro-4-((4,5- Dihydro-1H-Imidazol-2-Yl)Amino)Phenyl)-4-((4,5-Dihydro-1H-Imidazol-2- Yl)Amino)Benzamide] - (Drug JNI18)
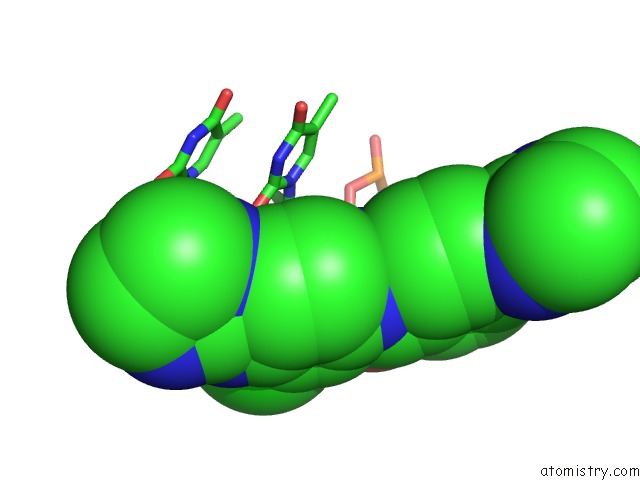
Mono view
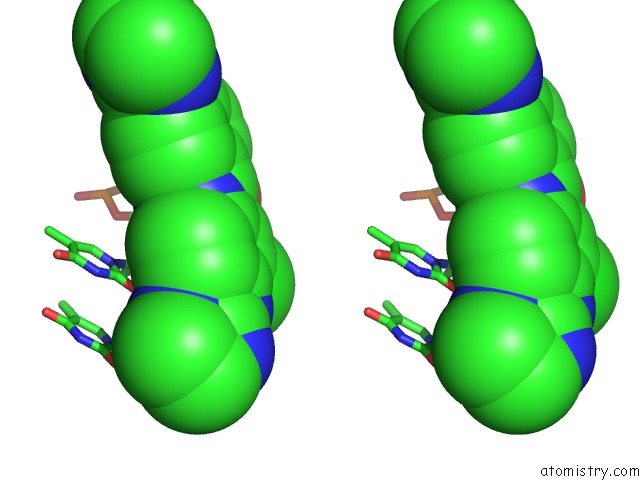
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Structure of the Dna Duplex D(Aaattt)2 with [N-(3-Chloro-4-((4,5- Dihydro-1H-Imidazol-2-Yl)Amino)Phenyl)-4-((4,5-Dihydro-1H-Imidazol-2- Yl)Amino)Benzamide] - (Drug JNI18) within 5.0Å range:
|
Chlorine binding site 2 out of 3 in 6gim
Go back to
Chlorine binding site 2 out
of 3 in the Structure of the Dna Duplex D(Aaattt)2 with [N-(3-Chloro-4-((4,5- Dihydro-1H-Imidazol-2-Yl)Amino)Phenyl)-4-((4,5-Dihydro-1H-Imidazol-2- Yl)Amino)Benzamide] - (Drug JNI18)
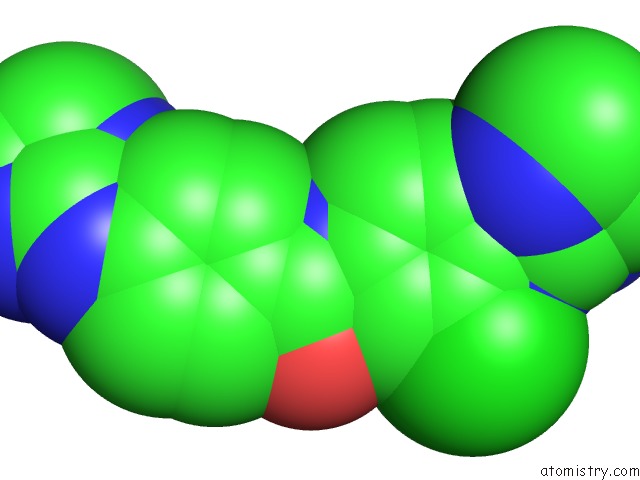
Mono view
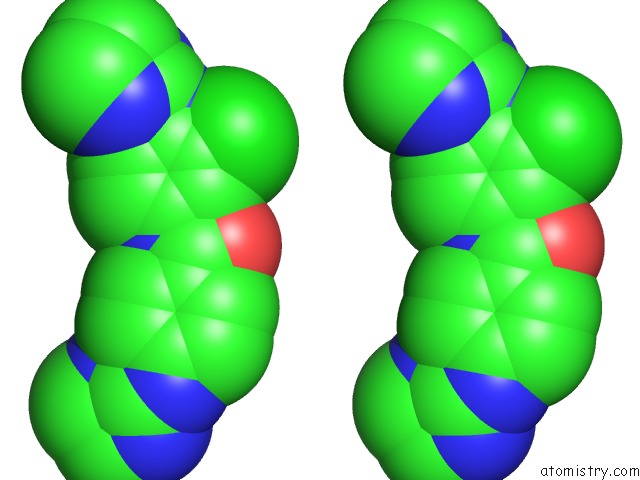
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Structure of the Dna Duplex D(Aaattt)2 with [N-(3-Chloro-4-((4,5- Dihydro-1H-Imidazol-2-Yl)Amino)Phenyl)-4-((4,5-Dihydro-1H-Imidazol-2- Yl)Amino)Benzamide] - (Drug JNI18) within 5.0Å range:
|
Chlorine binding site 3 out of 3 in 6gim
Go back to
Chlorine binding site 3 out
of 3 in the Structure of the Dna Duplex D(Aaattt)2 with [N-(3-Chloro-4-((4,5- Dihydro-1H-Imidazol-2-Yl)Amino)Phenyl)-4-((4,5-Dihydro-1H-Imidazol-2- Yl)Amino)Benzamide] - (Drug JNI18)
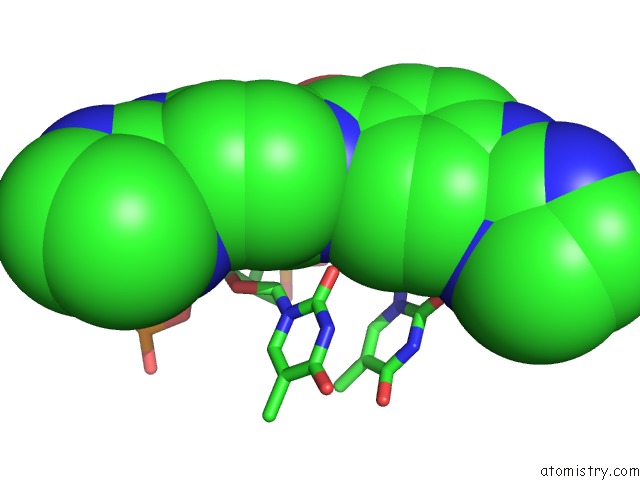
Mono view
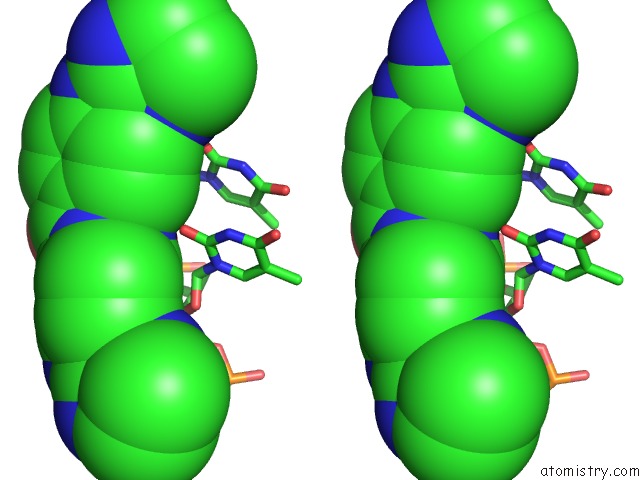
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Structure of the Dna Duplex D(Aaattt)2 with [N-(3-Chloro-4-((4,5- Dihydro-1H-Imidazol-2-Yl)Amino)Phenyl)-4-((4,5-Dihydro-1H-Imidazol-2- Yl)Amino)Benzamide] - (Drug JNI18) within 5.0Å range:
|
Reference:
C.R.Millan,
F.J.Acosta-Reyes,
L.Lagartera,
G.U.Ebiloma,
L.Lemgruber,
J.J.Nue Martinez,
N.Saperas,
C.Dardonville,
H.P.De Koning,
J.L.Campos.
Functional and Structural Analysis of at-Specific Minor Groove Binders That Disrupt Dna-Protein Interactions and Cause Disintegration of the Trypanosoma Brucei Kinetoplast. Nucleic Acids Res. V. 45 8378 2017.
ISSN: ESSN 1362-4962
PubMed: 28637278
DOI: 10.1093/NAR/GKX521
Page generated: Sat Jul 12 14:33:13 2025
ISSN: ESSN 1362-4962
PubMed: 28637278
DOI: 10.1093/NAR/GKX521
Last articles
F in 7PJRF in 7PKV
F in 7PKM
F in 7PKK
F in 7PK8
F in 7PJN
F in 7PK3
F in 7PJC
F in 7PJ2
F in 7PHN